WO1992014842A1 - Aptamers specific for thrombin and methods of use - Google Patents
Aptamers specific for thrombin and methods of use Download PDFInfo
- Publication number
- WO1992014842A1 WO1992014842A1 PCT/US1992/001367 US9201367W WO9214842A1 WO 1992014842 A1 WO1992014842 A1 WO 1992014842A1 US 9201367 W US9201367 W US 9201367W WO 9214842 A1 WO9214842 A1 WO 9214842A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- aptamer
- thrombin
- oligonucleotide
- oligonucleotides
- binding
- Prior art date
Links
Classifications
-
- G—PHYSICS
- G01—MEASURING; TESTING
- G01N—INVESTIGATING OR ANALYSING MATERIALS BY DETERMINING THEIR CHEMICAL OR PHYSICAL PROPERTIES
- G01N33/00—Investigating or analysing materials by specific methods not covered by groups G01N1/00 - G01N31/00
- G01N33/48—Biological material, e.g. blood, urine; Haemocytometers
- G01N33/50—Chemical analysis of biological material, e.g. blood, urine; Testing involving biospecific ligand binding methods; Immunological testing
- G01N33/86—Chemical analysis of biological material, e.g. blood, urine; Testing involving biospecific ligand binding methods; Immunological testing involving blood coagulating time or factors, or their receptors
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/11—DNA or RNA fragments; Modified forms thereof; Non-coding nucleic acids having a biological activity
- C12N15/111—General methods applicable to biologically active non-coding nucleic acids
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/11—DNA or RNA fragments; Modified forms thereof; Non-coding nucleic acids having a biological activity
- C12N15/115—Aptamers, i.e. nucleic acids binding a target molecule specifically and with high affinity without hybridising therewith ; Nucleic acids binding to non-nucleic acids, e.g. aptamers
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/10—Type of nucleic acid
- C12N2310/16—Aptamers
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/30—Chemical structure
- C12N2310/31—Chemical structure of the backbone
- C12N2310/315—Phosphorothioates
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/30—Chemical structure
- C12N2310/31—Chemical structure of the backbone
- C12N2310/318—Chemical structure of the backbone where the PO2 is completely replaced, e.g. MMI or formacetal
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/30—Chemical structure
- C12N2310/33—Chemical structure of the base
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/30—Chemical structure
- C12N2310/33—Chemical structure of the base
- C12N2310/332—Abasic residue
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/30—Chemical structure
- C12N2310/33—Chemical structure of the base
- C12N2310/335—Modified T or U
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2320/00—Applications; Uses
- C12N2320/10—Applications; Uses in screening processes
- C12N2320/13—Applications; Uses in screening processes in a process of directed evolution, e.g. SELEX, acquiring a new function
-
- G—PHYSICS
- G01—MEASURING; TESTING
- G01N—INVESTIGATING OR ANALYSING MATERIALS BY DETERMINING THEIR CHEMICAL OR PHYSICAL PROPERTIES
- G01N2333/00—Assays involving biological materials from specific organisms or of a specific nature
- G01N2333/90—Enzymes; Proenzymes
- G01N2333/914—Hydrolases (3)
- G01N2333/948—Hydrolases (3) acting on peptide bonds (3.4)
- G01N2333/974—Thrombin
Definitions
- This invention is in the field of rational drug design using biomolecule targeting and aptamer development.
- the invention discloses and claims methods for making aptamers to thrombin and the aptamers resulting therefrom which may be applied broadly to diagnostics and therapeutics. More specifically, this invention is related to aptamers that bind to thrombin and interfere with its normal biological function, and therapeutic uses for these aptamers.
- oligonucleotides have been termed "aptamers" herein.
- Tuerk and Gold describe the use of an in vitro selection and enrichment procedure. In this method, a pool of RNAs that are completely randomized at specific positions is subjected to selection for binding by a desired nucleic acid-binding protein which is then bound to a nitrocellulose filter. The bound RNAs then are recovered and amplified as double-stranded DNA that is competent for subsequent in vitro transcription.
- RNA binding region of T4 DNA polymerase The newly- transcribed RNA then is recycled through this procedure to enrich for oligonucleotides that have consensus sequences for binding by the cognate protein.
- the oligonucleotides so obtained then may be sequenced for further study. Tuerk and Gold applied this procedure to identify RNA oligonucleotides which are bound by the RNA binding region of T4 DNA polymerase.
- TDA target detection assay
- DNA aptamers 25 of single-stranded DNA as an appropriate material for generating aptamers.
- DNA aptamers has several advantages over RNA including increased nuclease stability (Shaw, J.P. et al., Nuc Acid Res (1991) 19:747- 750) , in particular plasma nuclease stability, and ease
- RNA generally is converted to DNA prior to amplification using reverse transcriptase, a process that is not equally efficient with all sequences, resulting in loss of some aptamers from a selected pool.
- Thrombin Acute vascular diseases are associated with partial or total occlusion of a blood vessel by blood clots, which contain platelets and fibrin. These diseases include serious health risks such as myocardial infarction, deep vein thrombosis, pulmonary embolism, peripheral arterial occlusion and the like. Treatment or prophylaxis of thrombotic diseases is based on either inhibition of clotting or acceleration of thrombolysis. Both approaches to treatment of thrombotic disease have been described using agents such as heparin or hirudin to inhibit thrombin and streptokinase or tissue plasminogen activator to accelerate thrombolysis.
- Thrombin is a multifunctional enzyme that (i) converts fibrinogen to fibrin by enzymatic cleavage; (ii) has mitogenic ef ects on lymphocytes and vascular smooth muscle cells; (iii) stimulates platelet aggregation and activation; (iv) is chemotactic for monocytes; (v) stimulates vascular endothelial cell mediated production of prostacyclin, platelet-activating factor and other factors; (vi) induces neutrophil adherence to vessel walls; (vii) stimulates vascular endothelial cell adhesion phenotype; and (viii) generates activated protein C by cleavage of protein C.
- Mitogenic activity of thrombin is exerted through binding to thrombin receptors (Coughlin, S.R., et al, J. Clin. Invest.. (1992) 81:351-355). Platelet aggregation, which plays a major role in arterial thrombosis is largely dependent on the function of thrombin (Hanson, S.R., et al, Proc. Natl. Acad. Sci. USA. (1988) £5_:3184-3188) . Platelets carry functional thrombin receptors. Inflammatory responses can also be mediated by thrombin through stimulation of platelet activating factor (PAF) (Prescott, S., et al, Proc. Natl. Acad. Sci. USA. (1984) £1:3534-3538. PAF promotes adhesion of neutrophils to endothelial matrix, leading to degranulation of the neutrophils and an associated inflammatory response.
- PAF platelet activating factor
- the thrombin aptamers bind to thrombin and inhibit both its catalytic activity in converting fibrinogen to fibrin and its platelet aggregating activity.
- the aptamers are potent inhibitors of thrombin function and represent a new cla* of pharmaceutical agents for modulation of the activity of this protease.
- the molecules of this invention may be utilized in compositions and methods for inhibiting any thrombin-mediated or thrombin-associated process or function.
- compositions containing these molecules as well as methods of treatment or prophylaxis of vascular diseases, inflammatory responses, cancer-related hypercoagulable states, sepsis and neural vasooclusive diseases using these compositions are also part of the present invention.
- These molecules can also be utilized in compositions and methods for in vitro or in vivo imaging, diagnosis, for storing and treating extracorporeal blood and for coating implant devices.
- the aptamers of the present invention are composed of DNA and chemically related molecules.
- DNA is a class of molecule ordinarily found in animals and it is expected that the immunogenicity of thrombin aptamers will be nonexistent or very low.
- Immune reactions against nucleic acids are known to be rare and, when observed, are associated with autoimmune disorders. Because of their compatibility with biological systems, the molecules of the invention are suitable in the treatment of both acute and nonacute vascular conditions.
- the invention is directed to a method to determine an aptamer which binds specifically to thrombin, which method comprises providing a mixture containing oligomers optionally having portions which form a random set of sequences and portions which permit amplification of the oligomers, incubating the oligomer mixture with thrombin coupled to a support to form complexes between thrombin and the oligomers bound specifically thereto, removing the unbound members of the oligonucleotide mixture from the support environment, recovering the complex ⁇ d oligonucleotide(s) from the support, amplifying the recovered oligonucleotides, and sequencing the recovered and amplified oligonucleotide(s; which had been complexed with thrombin.
- the starting mixture of oligonucleotides having random sequences may also contains a consensus sequence known to bind to thrombin.
- this invention is directed to single-stranded deoxyribonucleotides that bind specifically to thrombin. It has been heretofore thought that the three-dimensional structure of double- stranded DNA limited the structural diversity of the molecule. The inventors herein are unaware of any prior demonstration of structural diversity for single- or double-stranded DNA sufficient to provide the range of conformations necessary to provide aptamers to biomolecules. For example, known RNA structures, such as pseudoknots, have not been described for single-stranded DNA.
- the invention is directed to oligonucleotides which contain sequences identified by the above methods, and to oligonucleotide sequences which bind specifically to thrombin. In still another aspect, the invention is directed to complexes comprising the thrombin target substance and specifically bound oligomer.
- the invention is directed to oligomers which contain sequences that bind specifically to thrombin target substances and inhibit its normal biological function, and to the use of these oligomers in therapy, diagnostics, and purification procedures.
- this invention is directed to oligomers which contain sequences that bind specifically to thrombin and inhibits its normal biological function, and which also contain one or more modified bases, sugars, or sugar linkages, and to the use of these oligomers in therapy, diagnostics, and purification procedures.
- Figure 1 is a chart depicting thrombin aptamer consensus-related sequences.
- Figure 2 is a plot of in vivo thrombin 5 inhibition obtained from primates using a 15-mer aptamer.
- these aptamers can be used as a separation tool for retrieving or detecting thrombin.
- the aptamers function much like monoclonal antibodies in their specificity and usage.
- thrombin can be recovered in useful quantities.
- these aptamers can be t used in diagnosis by employing them in specific binding assays.
- the aptamers of the invention may be coupled to auxiliary substances that enhance or complement the function of the aptamer.
- auxiliary substances include, for example, labels such as radioisotopes, fluorescent labels, enzyme labels and the like; specific binding reagents such as antibodies, additional aptamer sequence, cell surface receptor ligands, receptors per se and the like; toxins such as diphtheria toxin, tetanus toxin or ricin; drugs such as antiinflammatory, antibiotic, or metabolic regulator pharmaceuticals, solid supports such as chromatographic or electrophoretic supports, and the like.
- Suitable techniques for coupling of aptamers to desired auxiliary substances are generally known for a variety of such auxiliary substances, and the specific nature of the coupling procedure will depend on the nature of the auxiliary substance chosen.
- Coupling may be direct covalent coupling or may involve the use of synthetic linkers such as those marketed by Pierce Chemical Co., Rockford, IL.
- oligonucleotides or “aptamers” refers to oligonucleotides having specific binding regions which are capable of forming complexes with thrombin in an environment wherein other substances in the same environment are not complexed to the oligonucleotide.
- the specificity of the binding is defined in terms of the comparative dissociation constants of the. aptamer for thrombin as compared to the dissociation constant with respect to the aptamer and other materials in the environment or unrelated molecules in general.
- the Kd for the aptamer with respect to thrombin will be 2-fold, preferably 5-fold, more preferably 10-fold less than Kd with respect to thrombin and the unrelated material or accompanying material in the environment. Even more preferably the Kd will be 50- fold less, more preferably 100-fold less, and more preferably 200-fold less.
- the binding affinity of the aptamers herein with respect to thrombin is defined in terms of Kd.
- the value of this dissociation constant can be determined directly by well-known methods, and can be computed even for complex mixtures by methods such as those, for example, set forth in Caceci, M. , et al., Byte (1984) 9_:340-362. It has been observed, however, that for some small oligonucleotides, direct determination of Kd is difficult, and can lead to misleadingly high results. Under these circumstances, a competitive binding assay for thrombin may be conducted with respect to substances known to bind thrombin. The value of the concentration at which 50% inhibition occurs (Ki) is, under ideal conditions, equivalent to Kd.
- Ki in no event can Ki be less than Kd.
- determination of Ki in the alternative, sets a maximal value for the value of Kd.
- measurement of Ki can conveniently be substituted to provide an upper limit for Kd.
- thrombin As specificity is defined, in terms of Kd as set forth above, excluded from the categories of unrelated materials and materials accompanying thrombin in its environment are those materials which are sufficiently related to thrombin to be immunologically crossreactive therewith, and materials which natively bind oligonucleotides of particular sequences such as nucleases, restriction enzymes, and the like.
- immunologically crossreactive is meant that antibodies raised with respect to thrombin crossreact under standard assay conditions with the candidate material. Generally, for antibodies to crossreact in standard assays, the binding affinities of the antibodies for crossreactive materials as compared to thrombin should be in the range of 5-fold to 100-fold, generally about 10-fold.
- aptamers which contain specific binding regions are specific with respect to unrelated materials and with respect to materials which do not normally bind such oligonucleotides such as nucleases and restriction enzymes.
- a minimum of approximately 6 nucleotides, preferably 10, and more preferably 14 or 15 nucleotides, are necessary to effect specific binding.
- Aptamers of sequences as short as 6 bases have been shown to specifically bind and inhibit thrombin.
- the only apparent limitations on the binding specificity of the thrombin/oligonucleotide couples of the invention concern sufficient sequence to be distinctive in the binding oligonucleotide and sufficient binding capacity of thrombin to obtain the necessary interaction.
- Oligonucleotides of sequences shorter than 10, e.g., 6 mers, are feasible if the approp * ate interaction can be obtained in the context of the environment in which the thrombin is placed. Thus, if there are few interferences by other materials, less specificity and less strength of binding may be required.
- aptamer refers in general to either an oligonucleotide of a single defined sequence or a mixture of said oligonucleotides, wherein the mixture retains the properties of binding specifically to thrombin.
- aptamer denotes both singular and plural sequences of oligonucleotides, as defined herein.
- the aptamers of the invention are specifically binding oligonucleotides, wherein "oligonucleotide” is as defined herein.
- oligonucleotides include not only those with conventional bases, sugar residues and internucleotide linkages, but also those which contain modifications of any or all of these three moieties.
- Single-stranded oligonucleotides refers to those oligonucleotides which contain a single covalently linked series of nucleotide residues.
- Oligonucleotides include RNA or DNA sequences of more than one nucleotide in either single chain or duplex form and specifically includes short sequences such as di ers and trimers, in either single chain or duplex form, which may be intermediates in the production of the specifically binding oligonucleotides.
- Oligomer is generic to polydeoxyribonucleotides (containing 2'-deoxy-D-ribose or modified forms thereof), i.e., DNA, to polyribonucleo- tides (containing D-ribose or modified forms thereof) , i.e., RNA, and to any other type of polynucleotide which is an N-glycoside or C-glycoside of a purine or pyrimidine base, or modified purine or pyrimidine base.
- the oligomers of the invention may be formed using conventional phosphodiester-linked nucleotides and synthesized using standard solid phase (or solution phase) oligonucleotide synthesis techniques, which are now commercially available.
- the oligomers of the invention may also contain one or more "substitute” linkages as is generally understood in the art. Some of these substitute linkages are non-polar and contribute to the desired ability of the oligomer to diffuse across membranes.
- substitute linkages are defined herein as conventional alternative linkages such as phosphorothioate or phosphoramidate, are synthesized as described in the generally available literature.
- Alternative linking groups include, but are not limited to embodiments wherein a moiety of the formula P(0)S, ("thioate”), P(S)S ("dithioate”) , P(0)NR' 2 , P(0)R', P(0)0R , CO, or CONR', wherein R' is H (or a salt) or alkyl (1-12C) and R D is alkyl (1-9C) is joined to adjacent nucleotides through -0- or -S-. Dithioate linkages are disclosed and claimed in commonly owned U.S. application no. 248,517.
- Substitute linkages that may be used in the oligomers disclosed herein also include nonphosphorous-based internucleotide linkages such as the 3' -thioformacetal (-S-CH 2 -0-), formacetal (-0-CH 2 -0-) and 3' -amine (-NH-CH 2 -CH 2 -) internucleotide linkages disclosed and claimed in commonly owned pending U.S. patent application serial nos. 690,786 and 763,130, both incorporated herein by reference.
- One or more substitute linkages may be utilized in the oligomers in order to further facilitate binding with complementary target nucleic acid sequences or to increase the stability of the oligomers toward nucleases, as well as to confer permeation ability. (Not all such linkages in the same oligomer need be identical.)
- nucleoside or “nucleotide” is similarly generic to ribonucleosides or ribonucleotides, deoxyribonucleosides or deoxyribonucleotides, or to any other nucleoside which is an N-glycoside or C-glycoside of a purine or pyrimidine base, or modified purine or pyrimidine base.
- the stereochemistry of the sugar carbons may be other than that of D-ribose in one or more residues.
- analogs where the ribose or deoxyribose moiety is replaced by an alternate structure such as the 6-membered morpholino ring described in U.S.
- Nucleoside and “nucleotide” include those moieties which contain not only the natively found purine and pyrimidine bases A, T, C, G and U, but also modified or analogous forms thereof. Modifications include alkylated purines or pyrimidines, acylated purines or pyrimidines, or other heterocycles. Such “analogous purines” and “analogous pyrimidines” are those generally known in the art, many of which are used as chemotherapeutic agents.
- An exemplary but not exhaustive list includes pseudoisocytosme, N 4,N4-ethanocytosine, 8- hydroxy-N -methyladenine, 4-acetylcytosine, *5- (carboxyhydroxylmethyl) uracil, 5-fluorouracil, 5-bromouracil, 5-carboxymethylaminomethyl-2-thiouracil, 5-carboxymethylaminomethyl uracil, dihydrouracil, inosine, N -iscpentenyl-adenine, 1-methyladenine, 1-methylpseudouracil, 1-methylguanine, 1-methylinosine, 2,2-dimethylguanine, 2-methyladenine, 2-methylguanine, 3- methylcytosine, 5-methylcytosine, N -methyladenine, 7- methylguanine, 5-methylaminomethyl uracil, 5-methoxy aminomethyl-2-thiouracil, beta-D-mannosylqueosine, 5'- me
- nucleotide residues which are abasic, i.e., devoid of a purine or a pyrimidine base may also be included in the aptamers of the invention and in the methods for their obtention.
- sugar residues in the oligonucleotides of the invention may also be other than conventional ribose and deoxyribose residues.
- substitution at the 2' -position of the furanose residue is particularly important.
- Aptamer oligonucleotides may contain analogous forms of ribose or deoxyribose sugars that are generally known in the art.
- An exemplary, but not exhaustive list includes 2' substituted sugars such as 2' -O-methyl-, 2'- 0-alkyl, 2'-0-allyl, 2'-S-alkyl, 2'-S-allyl, 2'-fluoro-, 2' -halo, or 2' -azido-ribose, carbocyclic sugar analogs, Qf-anomeric sugars, epimeric sugars such as arabinose, xyloses or lyxoses, pyranose sugars, furanose sugars, sedoheptuloses, acyclic analogs and abasic nucleoside analogs such as methyl riboside, ethyl riboside or propyl riboside.
- the method for preparing the aptamers of the invention involves incubating thrombin with a mixture of oligonucleotides under conditions wherein some but not all of the members of the oligonucleotide mixture form complexes with the thrombin.
- the resulting complexes are then separated from the uncomplexed members of the oligonucleotide mixture and the complexed members which constitute an aptamer (at this stage the aptamer generally being a population of a multiplicity of oligonucleotide sequences) is recovered from the complex and amplified.
- the resulting aptamer (mixture) may then be substituted for the starting mixture in repeated iterations of this series of steps.
- the aptamer may be used as obtained or may be sequenced and synthetic forms of the aptamer prepared.
- the oligonucleotides used as members of the starting mixture may be single-stranded or double- stranded DNA or RNA, or modified forms thereof.
- single-stranded DNA is preferred.
- the use of DNA eliminates the need for conversion of RNA aptamers to DNA by reverse transcriptase prior to PCR amplification. Furthermore, DNA is less susceptible to nuclease degradation than RNA.
- the oligonucleotides that bind to thrombin are separated from the rest of the mixture and recovered and amplified. Amplification may be conducted before or after separation from thrombin.
- the oligonucleotides are conveniently amplified by PCR to give a pool of DNA sequences.
- the PCR method is well known in the art and described in, e.g., U.S. Patent Nos. 4,683,195 and 4,683,202 and Saiki, R.K., et al. , Science (1988) 3£:487-491, and European patent applications 86302298.4, 86302299.2 and 87300203.4, as well as Methods in Enzvmologv (1987) 15_5_:335-350. If RNA is initially used, the amplified DNA sequences are transcribed into RNA.
- the recovered DNA or RNA, in the original single-stranded or duplex form, is then used in another round of selection and amplification. After three to six rounds of selection/amplification, oligomers that bind with an affinity in the mM to ⁇ M range can be obtained and affinities below the ⁇ M range are possible. PCR may also be performed in the presence of thrombin.
- amplification may be employed including standard cloning, ligase chain reaction, etc. (See e.g., Chu, et al., U.S. Patent No. 4,957,858).
- linkers may be attached to each side to facilitate cloning into standard vectors. Aptamers, either in single or double stranded form, may be cloned and recovered thereby providing an alternative amplification method.
- Amplified sequences can be applied to sequencing gels after any round to determine the nature of the aptamers being selected by thrombin. The entire process then may be repeated using the recovered and amplified duplex if sufficient resolution is not obtained.
- Amplified sequences can be cloned and individual oligonucleotides then sequenced. The entire process can then be repeated using the recovered and amplified oligomers as needed.
- an aptamer that binds specifically to thrombin may be recovered as DNA or RNA in single-stranded or duplex form using conventional techniques.
- a selected aptamer may be sequenced and resynthesized using one or more modified bases, sugars and linkages using conventional techniques.
- the specifically binding oligonucleotides need to contain the sequence-conferring specificity, but may be extended with flanking regions and otherwise derivatized.
- the starting mixture of oligonucleotide may be of undetermined sequence or may preferably contain a randomized portion, generally including from about 3 to about 400 nucleotides, more preferably 10 to 100 nucleotides.
- the randomization may be complete, or there may be a preponderance of certain sequences in the mixture, or a preponderance of certain residues at particular positions.
- the randomized sequence is preferably flanked by primer sequences which permit the application of the polymerase chain reaction directly to the recovered oligonucleotide from the complex.
- the flanking sequences may also contain other convenient features, such as restriction sites which permit the cloning of the amplified sequence.
- These primer hybridization regions generally contain 10 to 30, more preferably 15 to 25, and most preferably 18 to 20, bases of known sequence.
- the oligonucleotides of the starting mixture may be conventional oligonucleotides, most preferably single-stranded DNA, or may be modified forms of these conventional oligomers as described hereinabove.
- standard oligonucleotide synthesis techniques may be employed.
- Oligonucleotides may also be synthesized using solution phase methods such as triester synthesis, known in the art. The nature of the mixture is determined by the manner of the conduct of synthesis. Randomization can be achieved, if desired, by supplying mixtures of nucleotides for the positions at which randomization is desired.
- nucleotides and any desired number of such nucleotides can be supplied at any particular step.
- any degree of randomization may be employed.
- Some positions may be randomized by mixtures of only two or three bases rather than the conventional four. Randomized positions may alternate with those which have been specified. It may be helpful if some portions of the candidate randomized sequence are in fact known.
- the starting mixture of oligonucleotides subjected to the invention method will have a binding affinity for thrombin characterized by a Kd of 1 ⁇ M or greater. Binding affinities of the original mixture for thrombin may range from about 100 ⁇ M to 10 ⁇ M to 1 ⁇ M but, of course, the smaller the value of the dissociation constant, the more initial affinity there is in the starting material for thrombin. This may or may not be advantageous as specificity may be sacrificed by starting the procedure with materials with high binding affinity.
- a ratio of binding affinity reflects the ratio of Kds of the comparative complexes. Even more preferred in the conduct of the method of the invention is the achievement of an enhancement of an affinity of a factor of 500 or more.
- the method of the invention can be conducted to obtain the invention aptamers wherein the 5 aptamers are characterized by consisting of single- stranded DNA, by having a binding affinity for thrombin represented by a Kd of 100 x 10 or less, by having a specificity representing by a factor of at least 2, and preferably 5, more preferably 10 with respect to 0 unrelated molecules, by having a binding region of less than 15 nucleotide residues or a total size of less than 16 nucleotide residues, or by binding to thrombin.
- the invention processes are also characterized by accommodating starting mixtures of oligonucleotides 5 having a binding affinity for thrombin characterized by a Kd of l ⁇ M or more by an enhancement of binding affinity of 50 or more, and by being conducted under physiological conditions.
- physiological conditions means 0 the salt concentration and ionic strength in an aqueous solution which characterize fluids found in human metabolism commonly referred to as physiological buffer or physiological saline. In general, these are represented by an intracellular pH of 7.1 and salt 5 concentrations Na + :5-15 mM, K + :140 mM, Mg +2 :0.3 mM,
- the use of physiological conditions in the C aptamer selection method is extremely important, particularly with respect to those aptamers that may be intended for therapeutic use.
- .As is understood in the art, the concentration of various ions, in particular, the ionic strength, and the pH value impact on the value of the dissociation constant of the thrombin/aptamer complex.
- the initial mixture of candidate oligonucleotides will include oligc. ars which contain at least one modified nucleotide residue or linking group.
- any modified nucleotides such as the presence of specific affinity agents in the purification of the desired materials.
- the modification In order for the modified oligomer to yield useful results, the modification must result in a residue which is "read" in a known way by the polymerizing enzyme used in the amplification procedure. It is not necessary that the modified residue be incorporated into the oligomers in the amplification process, as long it is possible to discern from the nucleotide incorporated at the corresponding position the nature of the modification contained in the candidate, and provided only one round of complexation/amplification is needed.
- modified residues of the invention are also susceptible to enzymatic incorporation into oligonucleotides by the commonly used polymerase enzymes and the resulting oligomers will then directly read on the nature of the candidate actually contained in the initial complex. It should be noted that if more than one round of complexation is needed, the amplified sequence must include the modified residue, unless the entire pool is sequenced and resynthesized to include the modified residue.
- modifications can be made to the base residues in a oligonucleotide sequence without impairing the function of polymerizing enzymes to recognize the modified base in the template or to incorporate the modified residue.
- modifications include alkylation of the 5-position of uridine, deoxyuridine, cytidine and deoxycytidine; the N -position of cytidine and deoxycytidine; the N -position of adenine and deoxyadenine; the 7-position of 7-deazaguanine, 7- deazadeoxyguanine, 7-deazaadenine and 7-deazadeoxy- adenine.
- the modified base may be included in the oligomeric mixtures useful in the method of the invention.
- sugar moiety may also be modified without affecting the capacity of the sequence to be usable as a specific template in the synthesis of new DNA or RNA.
- the efficacy of the process of selection and amplification depends on the ability of the PCR reaction faithfully to reproduce the sequence actually complexed to thrombin.
- the oligonucleotide contains modified forms of cytosine (C*)
- the PCR reaction must recognize this as a modified cytosine and yield an oligomer in the cloned and sequenced product which reflect this characterization.
- the modified form of cytosine (C*) is included in the PCR reaction as dC*TP, the resulting mixture will contain C* at positions represented by this residue in the original member of the candidate mixture.
- modified oligonucleotides and linking groups may arbitrarily be used in the synthesized form of the aptamer.
- modified oligonucleotides in the methods and aptamers of the invention provides a tool for expansion of the repertoire of candidates to include large numbers of additional oligonucleotide sequences.
- Such expansion of the candidate pool may be especially important as the demonstration of binding to proteins, for example, in the prior art is limited to those proteins known to have the capability to bind DNA.
- Modifications of the oligonucleotide may be necessary to include all desired sequences among those for which specific binding can be achieved.
- one preferred method comprises incubating thrombin with a mixture of oligonucleotides, wherein these oligonucleotides contain at least one modified nucleotide residue or linkage, under conditions wherein complexation occurs with some but not all members of the mixture; separating the complexed from uncomplexed oligonucleotides, recovering and amplifying the complexed oligonucleotides and optionally determining the sequence of the recovered nucleotides.
- amplification is also conducted in the presence of modified nucleotides.
- thrombin will be incubated with the starting mixture of oligonucleotides and, as usual, the complexes form separated from uncomplexed oligonucleotides.
- the complex oligonucleotides, which are now an aptamer, are recovered and amplified from the complex.
- the recovered aptamer is then mixed with the second undesired substance from which thrombin is to be distinguished under conditions wherein members of the aptamer population which bind to said second substance can be complexed. This complex is then separated from the remaining oligonucleotides of the aptamer. The resulting second uncomplexed aptamer population is then recovered and amplified. The second aptamer population is highly specific for thrombin as compared to the second substance.
- the negative selection step may be conducted first, thus mixing the original oligonucleotide mixture with the undesired substance to complex away the members of the oligonucleotide mixture which bind to the second substance; the uncomplexed oligonucleotides are then recovered and amplified and incubated with thrombin under conditions wherein those members of the oligonucleotide mixture which bind thrombin are complexed. The resulting complexes then removed from the uncomplexed oligonucleotides and the bound aptamer population is recovered and amplified as usual.
- the original oligonucleotide mixture can be synthesized according to the desired contents of the mixture and can be separated by adding the oligonucleotide mixture to a column containing covalently attached thrombin (see, Ellington, A.D., et al., Nature (1990) 346:818-822) or to thrombin in solution (see Blackwell et al., Science (1990) 25_0:1104-1110; Blackwell et al., Science (1990) 250:1149- 1151; or to thrombin bound to a filter (see Tuerk, C. , and Gold, L. , Science (1990) 249:505-510) .
- Complexes between the aptamer and thrombin are separated from uncomplexed aptamers using any suitable technique, depending on the method used for complexation. For example, if columns are used, non-binding species are simply washed from the column using an appropriate buffer. Specifically b ⁇ und material can then be eluted.
- the complexes can be separated from the uncomplexed oligonucleotides using, for example, the mobility shift in electrophoresis technique (EMSA) , described in Davis, R.L., et al., Cell (1990) 6J1:733.
- MSA mobility shift in electrophoresis technique
- aptamer-thrombin complexes are run on a gel and aptamers removed from the region of the gel where thrombin runs. Unbound oligomers migrate outside these regions and are separated away.
- unbound aptamers are eluted using standard techniques and the desired aptamer recovered from the filters.
- separation of the complexes involves detachment of thrombin-aptamer complexes from column matrices as follows.
- a column or other support matrix having covalently or noncovalently coupled thrombin is synthesized. Any standard coupling reagent or procedure may be utilized, depending on the nature of the support.
- covalent binding may include the formation of disulfide, ether, ester or amide linkages.
- the length of the linkers used may be varied by conventional means.
- Noncovalent linkages include antibody-antigen interactions, protein-sugar interactions, as between, for example, a lectin column and a naturally-occurring oligosaccharide unit on a peptide.
- Lectin columns are particularly suited for selecting thrombin aptamers.
- Lectins are proteins or glycoproteins that can bind to complex carbohydrates or oligosaccharide units on glycoproteins, and are well- described in The Lectins (I.E. Liener et al., eds.. Academic Press 1986) .
- Lectins are isolated from a wide variety of natural sources, including peas, beans, lentils, pokeweed and snails.
- Concanavalin A is a particularly useful lectin.
- linking chemistries are also available.
- disulfide-derivatized biotin Pierce
- the resulting thrombin-S-S-biotin complex could then be used in combination with avidin- derivatized support.
- Oligonucleotide-thrombin complexes could then be recovered by disulfide bond cleavage.
- Linking chemistries will be selected on the basis of (i) conditions or reagents necessary for maintaining the structure or activity of thrombin.
- the oligomer mixture is added to and incubated with the support to permit oligonucleotide-thrombin complexation.
- Complexes between the oligonucleotides and thrombin are separated from uncomplexed oligonucleotides by removing unbound oligomers from the support environment. For example, if columns are used, nonbinding species are simply washed from the column using an appropriate buffer. Following removal of unbound oligomers, the thrombin is uncoupled from the support. The uncoupling procedure depends on the nature of the coupling, as described above.
- Thrombin bound through disulfide linkages may be removed by adding a sulfhydryl reagent such as dithiothreitol or ⁇ - mercaptoethanol.
- Thrombin bound to lectin supports may be removed by adding a complementary monosaccharide (e.g., ⁇ -methyl-mannoside, N-acetyl glucosamine, glucose, N-acetyl galactosamine, galactose or other saccharides for concanavalin A) .
- Oligonucleotides specifically bound to thrombin can then be recovered by standard denaturation techniques such as phenol extraction.
- the method of elution of thrombin- oligonucleotide complex from a support has superior unexpected properties when compared with standard oligonucleotide elution techniques.
- This invention is not dependent on the mechanism by which these superior properties occur. However, without wishing to be limited by any one mechanism, the following explanation is offered as to how more efficient elution is obtained.
- Certain support effects result from the binding of oligonucleotides to the support, or the support in conjunction with oligonucleotide or thrombin.
- Removing oligonucleotide-thrombin complexes enables the recovery of oligonucleotides specific to thrombin only, while eliminating oligonucleotides binding to the support, or the support in conjunction with oligonucleotide or thrombin.
- this method may give up to 1,000-fold enrichment for specifically binding species. Selection with thrombin remaining bound to support gives less enrichment per cycle, making it necessary to go through many more cycles in order to get a good aptamer population.
- Aptamers can also be selected in the above methods using a pool of oligonucleotides that vary in length as the starting material.
- a pool of oligonucleotides that vary in length as the starting material.
- several pools of oligonucleotides having random sequences are synthesized that vary in length from e.g. 50 to 60 bases for each pool and containing the same flanking primer-binding sequences.
- Equal molar amounts of each pool are mixed and the variable-length pool is then used to select for aptamers that bind to thrombin, as described above.
- This protocol selects for the optimal species for thrombin binding from the starting pool and does not limit aptamers to those of a given length.
- pools of mixed length aptamers can be used in parallel in separate selections and then combined and further selected to obtain the optimal binders from the size range initially used.
- three pools, A, B and C can be used.
- Pool A can consist of oligonucleotides having random sequences that vary in length from e.g. 30 to 40 bases;
- pool B can have sequences varying in length from e.g. 40 to 50 bases;
- pool C can have sequences varying in length from 50 to 60 bases. It is to be understood that the lengths described above are for illustrative purposes only. After selection to obtain binders from A, B, and C, all aptamers are mixed together.
- a number of rounds of selection are done as described above to obtain the best binders from the initial species selected in the 30- to 60-base range. Note that with this technique, not all possible species in some of the pools are used for selection. If the number of sites available for binding are increased, i.e., if a column is used and the size of the column increased, more species can be included for selection. Furthermore, this method allows for the selection of oligomers from the initial starting pool that are of optimal length for binding thrombin.
- Aptamers containing the specific binding sequences discerned through the method of the invention can also be derivatized in various ways. For example, if the aptamer is to be used for separation of thrombin, conventionally the oligonucleotide will be derivatized to a solid support to permit chromatographic separation. If the oligonucleotide is to be used for attaching a detectable moiety to thrombin, the oligonucleotide will be derivatized to include a radionuclide, a fluorescent molecule, a chromophore or the like. If the oligonucleo ⁇ tide is to be used in specific binding assays, coupling to solid support or detectable label, and the like are also desirable.
- the oligonucleotide may be derivatized to include ligands which permit easier transit of cellular barriers, toxic moieties which aid in the therapeutic effect, or enzymatic activities which perform desired functions at the thrombin site.
- the aptamer may also be included in a suitable expression system to provide for in situ generation of the desired sequence.
- Consensus sequences refers to a nucleotide sequence or region (which may or may not be made up of contiguous nucleotides) , which is found in one or more regions of at least two aptamers, the presence of which may be correlated with aptamer-to-thrombin-binding or with aptamer structure.
- a consensus sequence may be as short as three nucleotides long.
- Consensus sequences may be identified by sequence comparisons between individual aptamer species, which comparisons may be aided by computer programs and other tools for modeling secondary and tertiary structure from sequence information.
- the consensus sequence will contain at least 5 about 3 to 20 nucleotides, more commonly from 6 to 10 nucleotides.
- Consensus sequence means that certain positions, not necessarily contiguous, of an oligonucleotide are specified. By specified is meant 0 that the composition of the position is other than completely random. Not all oligonucleotides in a mixture may have the same nucleotide at such position; for example, the consensus sequence may contain a known ratio of particular nucleotides.
- a consensus 5 sequence might consist of a series of four positions wherein the first position in all members of the mixture is A, the second position is 25% A, 35% T and 40% C, the third position is T in all oligonucleotides, and the fourth position is G in 50% of the oligonucleotides and C 0 in 50% of the oligonucleotides.
- oligonucleotides that contain that sequence may be made by conventional synthetic or recombinant means. These aptamers, termed “secondary aptamers,” may also function 5 as thrombin-specific aptamers of this invention.
- a secondary aptamer may conserve the entire nucleotide sequence of an isolated aptamer, or may contain one or more additions, deletions or substitutions in the nucleotide sequence, as long as a consensus sequence is C conserved.
- a mixture of secondary aptamers may also function as thrombin-specific aptamers, wherein the mixture is a set of aptamers with a portion or portions of their nucleotide sequence being random or varying, and a conserved region which contains the consensus sequence. Additionally, secondary aptamers may be synthesized using one or more of the modified bases, sugars and linkages described herein using conventional techniques and those described herein.
- the aptamers of the invention are useful in diagnostic, research and therapeutic contexts.
- the thrombin aptamers have in vivo and ____: vivo clinical utilities, as indicated above.
- the aptamers may be used in the treatment or prevention of (i) restenosis or myointimal thickening associated with angioplasty, (ii) accelerated atherosclerosis after heart transplant operations, (iii) vascular graft reocclusion associated with vascular shunt implants, (iv) clotting or thrombus formation at the site of indwelling arterial or venous access lines, (v) thrombus formation associated with cardiopulmonary bypass surgery, (vi) thrombus formation associated with extracorporeal circuits that are used during various ex vivo procedures such as blood dialysis or apheresis, (vii) sepsis-related disseminated intravascular coagulation and (viii) coagulation in patients with known heparin allergy
- these aptamers are well suited for binding to biomolecules that are identical or similar between different species, where standard antibodies may be difficult to obtain. They are also useful in inhibition assays when the aptamers are chosen to inhibit the biological activity of thrombin.
- Antibodies are generally used to bind analytes that are detected or quantitated in various diagnostic assays. Aptamers represent a class of molecules that may be used in place of antibodies for in vitro or .in vivo diagnostic and purification purposes. Aptamers that bind to thrombin may be used as in vivo imaging or diagnostic reagents when suitably radiolabeled.
- Isotopes such as 131 I, 99m Tc, 90 Y, l ⁇ :L In and J "" ⁇ have been used to label various proteins or antibodies as is described in the literature (Cohn, K.H., et al, .Arch. Surg. (1987) 122.:1245-1429; Baidoo, K.E., et al, Cancer Res. (Suppl.) (1990) 5_0_:799s-803s; Beatty, J.D., et al, Cancer Res. (Suppl.) (1990) 50:840s-845: Sharkey, R.M., et al Cancer Res. (1988) 18:32270-3275).
- a preferred isotope is 99m Tc which is utilized as described in the literature.
- Chemical modifications of oligonucleotides that are compatible with labeling protocols are also known in the art and have been extensively described (Uhlmann, E., et al, Chemical Rev. (1990) 9_0:543-584; international publication Nos. WO 91/14696 and WO 91/13080) .
- the thrombin aptamers may also be labelled by linking a moiety that chelates an imaging agent such as 99 ⁇ Tc.
- an imaging agent such as 99 ⁇ Tc.
- thrombin aptamer would be administered to a patient followed by administration of the imaging agent. In vivo chelation of the imaging agent would occur, allowing subsequent imaging by conventional means.
- Thrombin aptamers may also be labeled with contrast agents such as lanthanide or transition metal complexes or nuclei such as 19 F, 15 N or 32 P to facilitate in vivo imaging of clots and similar formations. Imaging would be performed using magnetic resonance imaging techniques known in the art.
- contrast agents such as lanthanide or transition metal complexes or nuclei such as 19 F, 15 N or 32 P to facilitate in vivo imaging of clots and similar formations. Imaging would be performed using magnetic resonance imaging techniques known in the art.
- One consideration in generating radiolabeled antibodies is that the labeling procedure must not destroy its antigen-binding properties. This usually requires an optimized protocol to be generated for each isotope and antibody. Because the aptamers of the invention are tolerant of harsh chemical conditions, including conditions under which they are synthesized, facile radiolabeling of thrombin aptamers can be conducted without regard to loss of aptamer structure. Only the chemical integrity of the aptamer molecule must be preserved.
- the aptamers of the invention can be denatured without loss of their capacity to bind thrombin once placed under physiological conditions. Antibodies cannot be reversibly denatured in this manner.
- MAbs monoclonal antibodies
- Another consideration relevant to the use of monoclonal antibodies (MAbs) for in vivo imaging is their antigenicity. MAbs are usually derived from mouse hybridomas and as such are foreign proteins. When used in humans they elicit immune responses that limits their use in individual patients to one or two exposures. Once immunized, anti-MAb antibodies generated by an immunized individual leads to rapid clearance of the MAb. This consideration is also relevant to "humanized" MAbs that contain both mouse and human protein sequences.
- the aptamers described herein have a short half-life, a property that can permit rapid in vivo imaging after administration of labeled compound.
- the thrombin aptamers can also be advantageously used to avoid anaphylactic reactions such as those associated with imaging procedures that use conventional ionic or nonionic contrast agents.
- the aptamers also have a low molecular weight compared to .Abs, which can facilitate their penetration of a target structure, such as a clot, for imaging purposes.
- Radiolabeled thrombin aptamers can be used to image arteries or veins according to various clinical indications.
- aptamers can be used after angioplasty to image clots, including deep vein clots, CNS thromboses, pulmonary emboli, brain thromboses and the like.
- the aptamers of the invention are therefore particularly useful as diagnostic reagents to detect the presence or absence of thrombin.
- In vitro diagnostic tests are conducted by contacting a sample with the specifically binding oligonucleotide to obtain a complex which is then detected by conventional means.
- the aptamers may be labeled using radioactive, fluorescent, or chromogenic labels and the presence of label bound to solid support to which the thrombin has been bound through a specific or nonspecific binding means detected.
- the specifically binding oligonucleotides may be used to effect initial complexation to the support. Means for conducting assays using such oligomers as specific binding partners will track those for standard specific binding partner based assays.
- the oligomers of the invention are characterized by their ability to bind thrombin regardless of the mechanisms of binding or the mechanism of the effect thereof.
- the specifically binding oligonucleotides of the invention are especially helpful in effecting the isolation and purification of substances to which they bind.
- the aptamer containing the specific binding sequences is conjugated to a solid support and used as an affinity ligand in chromatographic separation of thrombin.
- the aptamers of the invention can be formulated for a variety of modes of administration, including systemic and topical or localized administration. Techniques and formulations generally may be found in Remington's Pharmaceutical Sciences. Mack Publishing Co., Easton, PA, latest edition. In general, the dosage required for therapeutic efficacy will range from about 0.1 ⁇ g to 20 mg aptamer/kg body weight. Alternatively, dosages within these ranges can be administered by constant infusion over an extended period of time, usually exceeding 24 hours, until the desired therapeutic benefits have been obtained.
- the aptamers of the invention are formulated in liquid solu ⁇ tions, preferably in physiologically compatible buffers such as Hank's solution or Ringer's solution. In addi- tion, the aptamers may be formulated in solid form and redissolved or suspended immediately prior to use. Lyophilized forms are also included.
- Systemic administration can also be by transmucosal or transdermal means, or the oligomers can be administered orally. Additional formulations which are suitable for other modes of administration include suppositories, intranasal and other aerosols.
- penetrants appropriate to the barrier to be permeated are used in the formulation. Such penetrants are generally known in the art, and include, for example, for transmucosal administration bile salts and fusidic acid derivatives. In addition, detergents may be used to facilitate permeation.
- Transmucosal administration may be through nasal sprays, for example, or using suppositories.
- the oligomers are formulated into conventional oral administration forms such as capsules, tablets, and tonics.
- the oligomers of the invention are formulated into ointments, salves, gels, or creams, as is generally known in the art.
- the aptamers may also be employed in expression systems, which are administered according to techniques applicable, for instance, in applying gene therapy.
- DNA oligonucleotides containing a randomized sequence region were synthesized using standard solid phase techniques and phosphoramidite chemistry (Oligonucleotide Synthesis. Gait, M.J., ed. (IRL Press), 1984; Cocuzza, A., Tetrahedron Letters. (1989) 30.:6287- 6291) .
- a 1 ⁇ M small-scale synthesis yielded 60 nmole of HPLC-purified single-stranded randomized DNA.
- DNA 18-mers with the following sequences were used as primers for PCR amplification of oligonucleotide sequences recovered from selection columns.
- the 5' primer sequence was 5' HO-CGTACGGTCGACGCTAGC-OH 3' and the 3' primer sequence was 5' biotin-O-
- biotin residue was linked to the 5' end of the 3' primer using commercially available biotin phosphoramidite (New England Nuclear, Cat. No. NEF-707) .
- the biotin phosphoramidite is incorporated into the strand during solid phase DNA synthesis using standard synthesis conditions.
- DNA 19-mers with the following sequences can also be used as primers for PCR amplification of oligonucleotides recovered from selection columns.
- the 3' primer sequence is
- the duplex form of the primer binding sites contain restriction enzyme sites.
- a pool of aptamer DNA 96 bases in length was synthesized as described in Example 1-A, and then PCR- amplified to construct the initial pool.
- a small amount of the enzymatically-synthesized DNA was further amplified in the presence of ⁇ - 32P-dNTPs to generate labeled aptamer to permit quantitation from column fractions.
- a thrombin column was prepared by washing 1 mf
- Con-A agarose-bound concanavalin A
- Con-A Vector Laboratories, cat. no. AL-1003
- 20 mM Tris-acetate buffer pH 7.4
- 1 mM MgCl 2 1 mM CaCl 2
- 5 mM KC1 5 mM KC1
- 140 mM NaCl the "selection buffer”
- 1 mf of settled support was then incubated overnight at 4°C in 10 mf selection buffer containing 225 ⁇ g (6.25 nmole) thrombin (Sigma, Cat. no. T-6759) .
- a column containing 1 mf of settled beads had a void volume of approximately 300 ⁇ L.
- a control Con-A column was prepared by adding 1 mf of settled support to a column followed by 5 washes of 1 mf of selection buffer.
- the DNA was heated in selection buffer at 95°C for 3 minutes and then cooled on ice for 10 minutes.
- the pool consisting of 100 pmole DNA in 0.5 mf selection buffer, was then pre-run on the control Con-A column at room temperature to remove species that bound to the control support.
- Three additional 0.5 mf aliquots of selection buffer were added and column fractions 2, 3 and 4 (0.5 mf each) were pooled and then reapplied to the column twice.
- the DNA in 1.5 mf selection buffer was then recovered. Approximately 1% of total input cpm were retained on the column.
- the recovered DNA was then applied to a Con-A- thro bin column as a 0.5 mf aliquot followed by a 1.0 mf aliquot. Flow-through was retained and reapplied to the column twice. DNA added to the column on the final application was left on the column for 1 hour at room temperature. The column was then eluted with 0.5 mf aliquots of selection buffer. 0.5 mf fractions were collected and radioactivity was determined in each fraction. Radioactivity in eluted fractions 7 through 12 were low and relatively constant. After recovery of fraction 12, the column was washed with 0.5 mf aliquots of 0.1 M ⁇ -methyl-mannoside (Sigma Cat. no.
- Fractions 14 and 15 showed a significant peak of thrombin enzyme activity, as determined spectrophotometrically by conversion of a chromogenic substrate (Kabi Diagnostica, Cat. no. S-2238) . 0.01% of the input DNA eluted in these two fractions.
- Aptamer DNA (Round 1 DNA) was recovered from the thrombin by phenol extraction (2 x 0.5 m ) . The aqueous phase volume was reduced to about 250 ⁇ l by n- butanol extraction. Aptamer DNA was precipitated on dry ice using 3 volumes of ethanol and 20 ⁇ g of glycogen as a carrier. The DNA was pelleted, washed once in 70% ethanol and then dried.
- a 200 ⁇ l PCR reaction consisted of the following: 100 ⁇ l template 96- mer DNA (approximately 0.01 pmoles) ; 20 ⁇ l 10X buffer (100 mM Tris-Cl (pH 8.3), 500 mM KCl, 20 mM MgCl 2 ) ; 32 ⁇ l dNTP's (5 mM cone total, 1.25 mM each dATP, dCTP, dGTP, and dTTP) ; 20 ⁇ l primer 1 (biotinylated 18-mer, 50 ⁇ M) ; 20 ⁇ l primer 2 (18-mer, 50 ⁇ M) ; 6 ⁇ l ⁇ - 32 P-dNTP's (approximately 60 ⁇ Ci) ; and 2 ⁇ l Taq I Polymerase (10 units) .
- the reaction was covered with 2 drops NUJOL mineral oil.
- a control reaction was also performed without template aptamer
- 0.1 M ⁇ -methyl-mannoside in selection buffer was added as fraction 13 in each elution, and fractions 14 and 15 were retained and the DNA amplified. Due to slow leeching of thrombin from the column, DNA bound to thrombin could also be seen in earlier fractions in rounds 3-5.
- round 5 aptamer DNA was analyzed for specificity in a filter binding assay.
- nitrocellulose filters (1 cm diameter) prebound with salmon sperm DNA were used to bind either: (l) An unselected 96-mer oligonucleotide DNA pool, (2) unselected DNA with thrombin (60 pmole) , (3) Round 5 aptamer DNA and thrombin (60 pmole) , (4) Round 5 aptamer DNA alone, or (5) Round 5 aptamer DNA and ovalbumin (60 pmole) .
- 3.5 pmole of DNA was used and the 5 incubation was in 200 ⁇ L selection buffer at room temperature for 1 hour.
- the filters were then washed 3 times with 3.0 mf of selection buffer and radioactivity was counted to determine the amount of DNA that was retained as a thrombin complex. The results are shown in 0 Table 2.
- Unselected DNA did not show significant binding to the thrombin while selected aptamer DNA bound to ⁇ thrombin. Binding results show specific thrombin binding with no detectable ovalbumin binding.
- Example l-A The 36-mer primer was used to generate internal BamHl restriction sites to aid in cloning.
- the amplified Round 5 aptamer DNA was then cloned into pGEM 3Z (Promega) . 32 of the resulting clones were then amplified directly using the following 5' primer sequence:
- Filter binding assays using aptamer DNA from 14 of the clones were used to determine the dissociation constants (Kd) for thrombin as follows: Thrombin concentrations between 10 ⁇ M and 1 nM were incubated at room temperature in selection buffer for 5 minutes in the presence of 0.08 pmole of radiolabeled 96-mer derived from cloned Round 5 aptamer DNA. After incubation, the thrombin and aptamer mixture was applied to nitrocellulose filters (0.2 micron, 2.4 cm diameter) that were pretreated with salmon sperm DNA (1 mg/mf DNA in selection buffer) and washed twice with 1 mf selection buffer. After application of thrombin mixture, the filters were washed three times with 1 mf selection buffer The radioactivity retained on the filters was then determined. Kd values for the individual clones ranged from 50 to >2000 nM.
- the DNA sequence of the 60-nucleotide randomly- generated region from 32 clones was determined in order to examine both the heterogeneity of the selected population and to identify homologous sequences. Sequence analysis showed each of the 32 clones to be distinct. However, striking sequence conservation was found. The hexamer 5' GGTTGG 3' was found at a variable location within the random sequence in 31 of 32 clones, and five of the six nucleotides are strictly conserved in all 32. Additionally, in 28 of the 32 clones a second hexamer 5' GGNTGG 3', where N is usually T and never C, is observed within 2-5 nucleotides from the first hexamer.
- 28 clones contain the consensus sequence 5' GGNTGG(N) Z GGNTGG 3' where z is an integer from 2 to 5.
- the remaining 4 clones contain a "close variant sequence" (a sequence differing by only a single base) .
- a compilation of the homologous sequences are shown in Figure l. It should be noted that DNA sequencing of several clones from the unselected DNA population or from a population of aptamers selected for binding to a different target revealed no homology to the thrombin- selected aptamers. From these data we conclude that this consensus sequence contains a sequence which is responsible either wholly or in part, for conferring thrombin affinity to the aptamers.
- Clotting time for the thrombin-catalyzed conversion of fibrinogen (2.0 mg/mL in selection buffer) to fibrin at 37°C was measured using a precision coagulation timer apparatus (Becton-Dickinson, Cat. nos. 64015, 64019, 64020) .
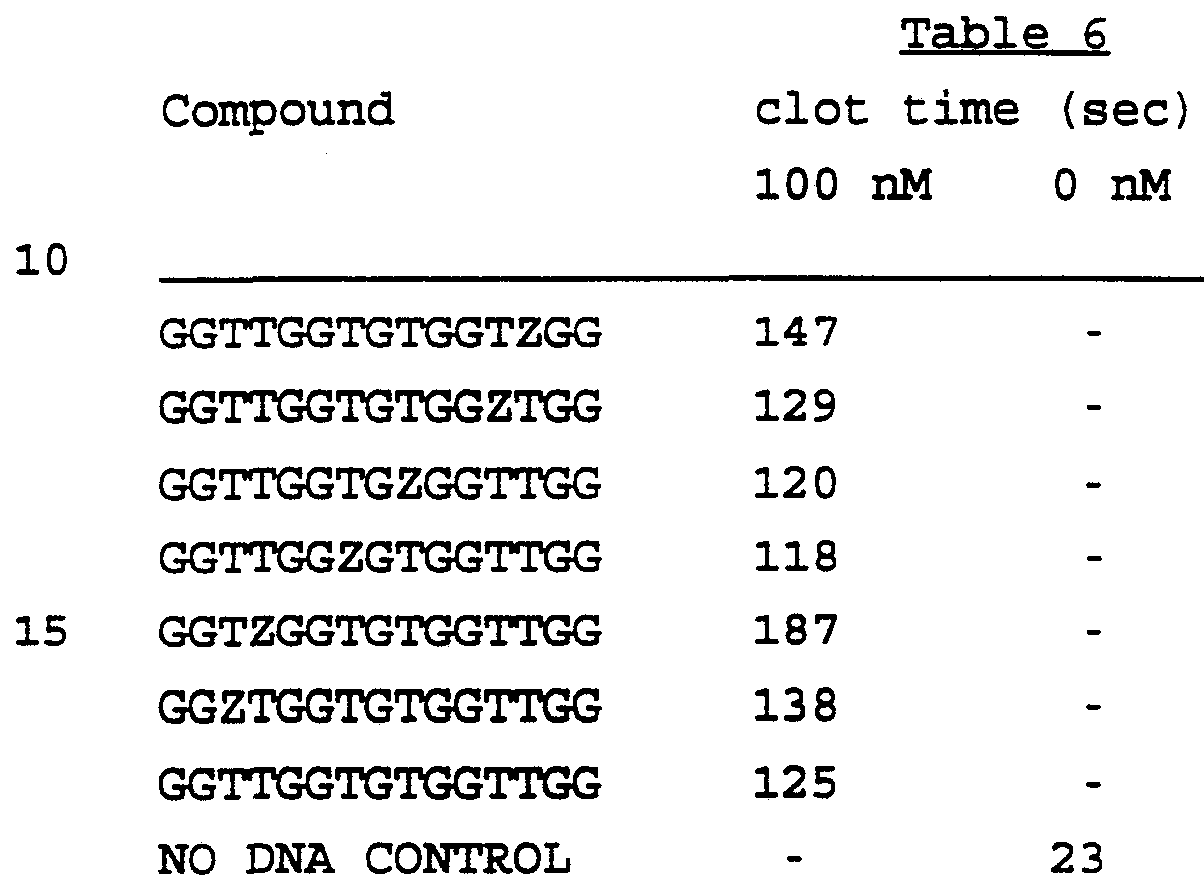
- Inhibition of thrombin activity was studied using a consensus-related sequence 7-mer, 5' GGTTGGG 3', or a control 7-mer with the same base composition but different sequence (5' GGGGGTT 3'). Clotting times were measured using the timer apparatus as above. The thrombin clotting time in this experiment was 24 sec using thrombin alone (10 nM) , 26 sec with thrombin and the control sequence at 20 ⁇ M and 38 sec with thrombin plus the consensus sequence at 20 ⁇ M, indicating specificity for thrombin inhibition at the level of the 7-mer.
- the inhibitory aptamers were active at physiological temperature under physiologic ion conditions and were able to bind to thrombin in the presence of the fibrinogen substrate, a key requirement for therapeutic efficacy.
- Example 2 Modified Thrombin Aptamers
- Thrombin Aptamers Modified forms of the single-stranded, thrombin consensus sequence-containing deoxynucleotide 15-mer described in Example 2, 5' GGTTGGTGTGGTTGG 3', and a closely related 17-mer, were synthesized using conventional techniques. These aptamers for the most part contain the identical nucleotide sequences, bases, sugars and phosphodiester linkages as conventional nucleic acids, but substitute one or more modified linking groups (thioate or MEA) , or modified bases (uracil or 5- (i-pentynyl-2' -deoxy)uracil) .
- modified linking groups thioate or MEA
- modified bases uracil or 5- (i-pentynyl-2' -deoxy)uracil
- the aptamers containing 5- (1-pentynyl) -2' -deoxyuridine were generated by replacing thymidine in the parent aptamers.
- Thrombin aptamers containing 5- (1-pentynyl) -2' -deoxyuridine were also obtained by selection as described in Examples 8 and 9 below.
- Example 3 Incorporation of 5- (l-pentynyl) -2' -deoxyuridine Into Aptamer
- Candidate DNA 5- (l-pentynyl) -2' -deoxyuridine was synthesized and converted to the triphosphate as described in Otvos, L., et al., Nucleic Acids Res (1987) 1763-1777.
- the pentynyl compound was obtained by reacting 5-iodo-2'- deoxyuridine with 1-pentyne in the presence of palladium catalyst.
- 5- (l-pentynyl) -2' -deoxyuridine triphosphate was then used as a replacement for thymidine triphosphate in the standard PCR reaction.
- a pool of 96-mer single-stranded DNA was synthesized, each strand consisting of specific 18-mer PCR primer sequences at both the 5' and 3' ends and a random 60-mer sequence in the center of the oligomer. Details of synthesis of the pool of single-stranded DNA is disclosed in Example 1 above. PCR conditions were the same as those described above, with the following changes. dATP, dGTP and dCTP were all used at a concentration of 200 ⁇ M. The optimal concentration for synthesis of full-length 96-mer DNA via PCR using 5- (1- pentynyl) -2' -deoxyuridine was 800 ⁇ M.
- 5-position of uridine, deoxyuridine, and at the N -position of cytidine and deoxycytidine are synthesized using methods described above. Each compound is converted to the triphosphate form and tested in the PCR assay described in Example 1 using an appropriate mixture of three normal deoxytriphosphates or ribotriphosphates along with a single modified base analog.
- Modified forms of the 15-mer thrombin aptamer, 5' GGTTGGTGTGGTTGG 3' containing one abasic residue at each position in the aptamer were synthesized and assayed for thrombin inhibition as described above.
- the abasic residue, l,4-anhydro-2-deoxy-D-ribitol was prepared as described in Eritja, R. , et al, Nucleosides and Nucleotides (1987) 6 :803-814.
- the N,N-diisopropylamino cyanoethylphosphoramidite synthon was prepared by standard methods as described in Caruthers, M.H. Accounts 5 Chem. Res.
- Example 8 Incorporation of 5- (1-pentvnyl) -2' -deoxyuridine Into Aptamer
- Candidate DNA 5- (l-pentynyl) -2' -deoxyuridine was synthesized and converted to the triphosphate as described in Otvos, L., et al., Nucleic Acids Res (1987) 1763-1777.
- the pentynyl compound was obtained by reacting 5-iodo-2'- deoxyuridine with 1-pentyne in the presence of a palladium catalyst.
- 5- (l-pentynyl) -2' -deoxyuridine triphosphate was then used as a replacement for thymidine triphosphate in the standard PCR reaction.
- a pool of 60-mer single-stranded DNA was synthesized, each strand consisting of specific 18-mer PCR primer sequences at both the 5' and 3' ends and a rando 20-mer sequence in the center of the oligomer. Details of synthesis of the pool of single-stranded DNA is disclosed in Example 1.
- VENTTM thermostable polymerase (New England Biolabs, Cat. No. 254) was employed. Amplification was performed as per the manufacturers instructions. Pentynyl dUTP was included in the reaction as a substitute for dTTP. The single-stranded 60-mer was isolated by a modification of standard procedures. The 200 ⁇ L PCR amplification reaction was divided into two samples which were applied to two NICKTM columns equilibrated (5 mL) as described. The eluent was collected, pooled and applied to avidin- agarose as described.
- the pool of aptamer DNA 60 bases in length was used essentially as described in Example 8.
- the aptamer pool sequence was 5' TAGAATACTCAAGCTTCGACG-N 20 -AGTTTGGATCCCCGGGTAC 3', while the 5' primer sequence was 5'TAGAATACTCAAGCTTCGACG 3' and the 3' biotin-linked primer was 5' GTACCCGGGGATCCAAACT 3' .
- Thrombin immobilized on a Con-A lectin column served as the target as described.
- aptamer DNA was recovered and amplified using thymidine triphosphate (dTTP) in place of 5- (l-pentynyl) -2' -deoxyuridine in order to facilitate subsequent cloning and replication of aptamer DNA in E__ coli.
- dTTP thymidine triphosphate
- the presence of a thymidine nucleotide at a given location in an aptamer corresponded to the location of a 5- (l-pentynyl) -2' - deoxyuridine nucleotide in each original round five aptamer.
- dTTP served to mark the location of 5- (l-pentynyl) -2' -deoxyuridine residues in the original selected DNA pools.
- the round five amplified DNA containing dTTP was digested with BamHI and Hindlll and cloned into the corresponding sites of pGEM 3Z (Promega Biotech) and transformed into E. coli.
- DNA from 21 clones was analyzed by dideoxy sequencing. Three of the clones contained aptamer sequences that were identical. Only one of the 21 clones contained a sequence that closely resembled the original 5' GGTTGG 3' binding motif obtained using thymine in the selection protocol.
- One of these two clones (#17) and the original unselected pool was analyzed for thrombin binding by nitrocellulose filter assay described above using DNA labeled with 32P to permi.t analysis of thrombi.n binding characteristics.
- the labeled DNA was synthesized by PCR and contained 5- (l-pentynyl) -2' -deoxyuridine in order to retain the original selected DNA structures.
- the DNA was incubated with thrombin at various concentrations between 10 nM and 10 ⁇ M to obtain the Kd values for thrombin binding.
- the Kd of the unselected pool was >10 ⁇ M while the Kd of clone 17 was 300 nM.
- Radiolabeled clone 17 DNA was synthesized using thymidine in place of 5- (l-pentynyl) -2' -deoxyuridine and the resulting DNA had a Kd of >10 ⁇ M, demonstrating that the 5- (l-pentynyl) -2'-deoxyuracil heterocycle could not be replaced by thymine in the selected aptamer without loss of binding affinity.
- 5 Representative sequences that were obtained are as follows.
- 5-methyl-2'-deoxycytidine triphosphate was obtained commercially (Pharmacia, Cat. No. 27-4225-01) and used to synthesize DNA containing random sequences 60 bases in length flanked by primers 19 bases in length.
- 3C The pool of aptamer DNA 98 bases in length was used essentially as described in Example 1. Thrombin immobilized on a Con-A lectin column ser-ved as the target as described.
- ⁇ 3C using: 10 mM Tris-HCl, pH 8.3 at 25° C, 1.5 mM MgCl , 5C mM NaCl and 200 ⁇ M of each of dATP, dGTP, dTTP and 5- methyl-2' -deoxycytidine triphosphate. 20 ⁇ Ci each of ⁇ - P-dATP and dGTP were added to label the DNA. 1 nmole of 5' and 3' primer were added followed by addition of 0.2 pmole of 98-mer template pool DNA. Amplification was initiated by addition of 2 ⁇ L (10 U) of Taq polymerase followed by sealing of the reaction with a mineral oil overlay. About 16 cycles of amplification were performed followed by a 10 minute final extension to complete all duplex synthesis.
- Amplified DNA was recovered (100 ⁇ L aqueous phase) , n-butanol extracted (650 ⁇ L) and applied to a Nick column prewashed with 5 mL of buffer containing 100 mM Tris-HCl pH 7.5 and 100 mM NaCl. Eluted DNA was applied to a 0.5 mL avidin-agarose column prewashed in the same buffer and washed until DNA loss from the column was ⁇ 1000 cpm. Single stranded DNA was eluted from the avidin column by washing with 0.15 N NaCl and the eluate was neutralized to pH 7.0 using glacial acetic acid.
- the 98-mer DNA was exchanged into selection buffer on a second Nick column and, after heat denaturation for 3 min at 95° C followed by cooling on ice for 10 min, used in aptamer selection on thrombin lectin columns. 1 mL thrombin columns were equilibrated in selection buffer prior to addition of single-stranded DNA. The single- stranded DNA was recirculated for three complete passes. Upon completion of the third pass the peak radioactive element was then applied to a 1 mL ConA/thrombin column (charged with 3 nmoles of thrombin) . Radioactive single- stranded 98-mer was applied three times to this matrix.
- the column was stoppered and allowed to stand for 1 hr. The column was then washed with selection buffer and 0.5 mL aliquot fractions collected. A total wash volume of 6 mL was employed. At this time, 0.1 M oc-methyl-mannoside in selection buffer was then added, followed by a 4 mL total volume wash. Thrombin enzymatic activity was detected via chromogenic substrate monitored by absorbance at 405 nm. Peak thrombin fractions were pooled, extracted with phenol, and the volume reduced by nBuOH extraction. 20 ⁇ g glycogen was added, the single-stranded 98-mer precipitated via ethanol addition and pelleted via centrifugation. The pelleted DNA was resuspended in water and used as a template for PCR amplification. This protocol was repeated to obtain a pool of DNA that resulted from 5 rounds of selection on thrombin columns.
- Double-stranded DNA was digested with EcoRI and HinDIII and cloned into pGEM3Z. Aptamers were then transformed into E. coli and analyzed by dideoxy sequencing. Round five aptamer pool DNA bound to thrombin with a Kd of approximately 300 nM.
- aptamer binding was demonstrated using 32P radiolabeled DNA and a series of proteins.
- 96-mer clone #29 having the partial sequence 5'CGGGGAGAGGTTGGTGTGGTTGGC.AATGGCTAGAGTAGTGAC GTTTTCGCGGTGAGGTCC 3' was used. The consensus sequence is shown underlined.
- a 21-mer aptamer, 5' GGTTGGGCTCGTTGGGTTGGG 3' was tested for inhibition of another fibrinogen-cleaving enzyme ancrod, which was obtained commercially (Sigma, Cat. No. A-5042) .
- 21-mer had a of Ki for thrombin of about 100 nM and its Kd was about 350 nM.
- Clone #29 had a Kd of about 200 nK for thrombin.
- the aptamer was shown to specifically bind to thrombin by a filter binding assay. Briefly, radiolabeled aptamer DNA at.about a concentration of about 1 nM was incubated with the indicated protein for several minutes at room temperature, followed by filtration of the aptamer-protein mixture through a nitrocellulose filter. The filter was washed with 3 mL of selection buffer and then radioactivity bound to the filters was determined as a % of input radioactivity. Results obtained are shown in Table 7. Binding data is shown for both unselected 96-mer DNA and for two separate experiments with clone #29 96-mer. All proteins were tested at about l ⁇ M concentration except human serum albumin which was used at 100 ⁇ M. The results that were obtained demonstrated that the 96-mer specifically bound to thrombin and had little affinity for most of the other proteins tested.
- the thrombin 21-mer ancrod assay was conducted as follows. Ancrod was suspended in sterile water at a concentration of 44 U/mL. 10 ⁇ L ancrod solution was added to 95 ⁇ L of selection buffer prewarmed to 37°C. 100 ⁇ L of this mixture was transferred to the coagulation cup of the fibrometer described above, followed by addition of 200 ⁇ L of fibrinogen and 20 ⁇ L of 21-mer DNA (both prewarmed to 37°C) . TE buffer pH 7.0 was used as a control lacking DNA.
- the control clot time was 25 seconds while the clot time in the presence of 500 nM 21- mer was 24 seconds and was 26 seconds in the presence of 33 ⁇ M 21-mer. This result demonstrated the specificity on inhibition of fibrinogen cleavage was limited to thrombin; ancrod was not affected.
- Example 12 Thrombin Aptamer Pharmacokinetic Studies A 15-mer single-stranded deoxynucleotide, 5' GGTTGGTGTGGTTGG 3', identified as a consensus sequence from 30 thrombin aptamer clones as described in Example 1 above, was used. Young adult rats of mixed gender and strain were used.
- the animals were anaesthetized and a diester of the 15-mer was injected through a catheter in 200 ⁇ l volumes (in 20 mM phosphate buffer, pH 7.4, 0.15 M NaCl) at two concentrations, so that the final concentration of 15-mer in the blood was about 0.5 and 5.0 ⁇ M respectively, although the exact concentration depends on the volume of distribution (which is unknown for this oligonucleotide) . These values are 10 to 100 times greater than the human in vitro Kd value. No heparin was used for catheterization.
- Example 13 Thrombin Aptamer Primate Studies Two thrombin aptamers were administered to adult male cynomologous monkeys. Unsubstituted 15-mer DNA with the sequence 5' GGTTGGTGTGGTTGG 3' and an analog, 5' GGTTGGTGTGGTT * G * G 3', containing thioate internucleotide linkages at the indicated positions (*) , were used. Aptamer was delivered as an intravenous bolus or infusion and then blood samples were withdrawn at various times after delivery of the bolus or during and after infusion. The catheter was heparinized after the 10 minute timepoint. The animals were not systematically heparinized.
- Thrombin inhibition was measured by a prothrombin time test (PT) using a commercially available kit, reagents and protocol (Sigma Diagnostics, St. Louis, catalog Nos. T 0263 and 870-3). Inhibition of thrombin was indicated by an increased clot time compared to the control in the PT test. Clot times were obtained by withdrawing a sample of blood, spinning out red cells and using the plasma in the PT test. Control thrombin PT clot time values were obtained several minutes prior to administration of aptamer.
- PT prothrombin time test
- the PT assay was conducted using 0.1 mL of monkey plasma prewarmed to 37° C and 0.2 mL of a 1:1 mixture of thromboplastin (used according to manufacturers instructions) and CaCl 2 (25 mM) , also prewarmed to 37°C. Thrombin clot times were measured with a fibrometer as described above.
- the animals were at least two years old and varied in weight from 4 to 6 kg. Doses of aptamer were adjusted for body weight. Aptamer DNA was dissolved in sterile 20 mM phosphate buffer (pH 7.4) at a concentration of 31.8 to 33.2 mg/mL and diluted in sterile physiological saline prior to delivery. Bolus injections were administered to give a final concentration of 22.5 mg/Kg (1 animal) of the diester aptamer or 11.25 mg/Kg (1 animal) of the diester aptamer.
- Infusions were administered over a 1 hour period to three groups of animals: (i) 0.5 mg/kg/min of diester 15-mer (4 animals), (ii) 0.1 mg/kg/min of diester 15-mer (2 animals) and (iii) 0.5 mg/kg/min of thioate analog 15- mer (2 animals) .
- PT assay results from the bolus injections showed thrombin inhibition times of 7.8, 3.3 and 1.35 times control at 2.5, 5.0 and 10.0 min respectively after delivery of the aptamer for the high dose animal. Inhibition times of 5.6, 2.2 and 1.2 times control were obtained from the low dose animal at the same time points.
- Figure 2 shows a plot of the PT times from the
- the data points show the PT clot time as an average value obtained from the 4 animals in the group.
- the arrows indicate time points at the beginning and end of the infusion period. Thrombin inhibition peaked at about 10 to 20 min after the infusion was initiated and remained level until the infusion period was terminated. Inhibitory activity decreased rapidly after the infusion of aptamer terminated.
- High dose diester and high dose thioate animals showed comparable inhibition of thrombin-mediated clotting, with the high dose thioate giving a sustained clot time of 2.5 to 2.7 times the control value during the course of the infusion.
- the low dose diester compound gave a clot time of 1.4 to 1.5 times the control value.
- Blood coagulation was marked by a pressure increase from about 50 mm Hg observed with uncoagulated blood (blood flow rate 200 mL/min) to pressure of at least 400 mm Hg.
- uncoagulated blood blood flow rate 200 mL/min
- pressure of at least 400 mm Hg was measured using citrated whole blood (recalcified at time zero) .
- citrated whole blood recalcified at time zero
- coagulation occurred at about 9 minutes after fresh blood was placed in the hemodialysis unit and circulation was begun.
- a heparin control (1 U/mL) gave sustained anticoagulation until the experiment was terminated at 80 minutes after start of circulation in the unit.
- Blood coagulation occurred at 51 minutes in one trial with the 15-mer. In a second trial, coagulation did not occur during the 80 minute course of the experiment.
Abstract
Oligonucleotide sequences that mediate specific binding to thrombin and optionally contain modified bases, sugars, or sugar linkages are disclosed. Single-stranded DNA oligomers are obtained that bind thrombin and inhibit its function in vitro and in vivo. The thrombin binding oligomers are useful for therapeutic, diagnostic and manufacturing purposes. An improved method for identifying these oligomers is also described, involving complexation of the support-bound thrombin with a mixture of oligonucleotides containing random sequences under conditions wherein a complex is formed with the specifically binding sequences, but not with the other members of the oligonucleotide mixture. The thrombin-oligonucleotide complexes are then separated from both the support and the uncomplexed oligonucleotides and the complexed members of the oligonucleotide mixture are recovered from the separated complex and subsequently amplified using standard techniques.
Description
APTAMERS SPECIFIC FOR THROMBIN AND METHODS OF USE
Technical Field
This invention is in the field of rational drug design using biomolecule targeting and aptamer development. The invention discloses and claims methods for making aptamers to thrombin and the aptamers resulting therefrom which may be applied broadly to diagnostics and therapeutics. More specifically, this invention is related to aptamers that bind to thrombin and interfere with its normal biological function, and therapeutic uses for these aptamers.
Background and Related Art
Specifically Bindinσ Oliσonucleotides. Con¬ ventional methods of detection and isolation of proteins and other molecules have employed antibodies and the like which specifically bind such substances. Recently, however, the cje novo design of specifically binding oligonucleotides for non-oligonucleotide targets that generally bind nucleic acids has been described. See, e.g., Blackwell, T.K., et al., Science (1990) 25_0:1104- 1110; Blackwell, T.K. , et al., Science (1990) 250:1149- 1152; Tuerk, C. , and Gold, L., Science (1990) 249:505- 510; Joyce, G.F., Gene (1989) 82.:83-87. Such oligonucleotides have been termed "aptamers" herein. Tuerk and Gold describe the use of an in vitro selection and enrichment procedure. In this method, a pool of RNAs that are completely randomized at specific positions is subjected to selection for binding by a desired nucleic acid-binding protein which is then bound to a nitrocellulose filter. The bound RNAs then are recovered and amplified as double-stranded DNA that is competent
for subsequent in vitro transcription. The newly- transcribed RNA then is recycled through this procedure to enrich for oligonucleotides that have consensus sequences for binding by the cognate protein. The oligonucleotides so obtained then may be sequenced for further study. Tuerk and Gold applied this procedure to identify RNA oligonucleotides which are bound by the RNA binding region of T4 DNA polymerase.
Kinzler, K. ., et al., Nucleic Acids Res. (1989) 17:3645-3653, applied this technique to identify double-stranded DNA sequences that were bound by proteins that bind to DNA .and regulate gene expression. In the reported work, total genomic DNA is first converted to a form that is suitable for amplification by PCR by ligation of linker sequences to the genomic DNA fragments and the DNA sequences of interest are selected by binding mediated by the target regulatory protein. The recovered bound sequences are then amplified by PCR. The process of binding by protein and amplification are repeated as needed. The selection and amplification process are repeated as needed. The process as described was applied to identify DNA sequences which bind to the Xenopus laevis transcription factor 3A. The same authors (Kinzler et al.) in a later paper, Mol. Cell Biol. (1990) 10.:634-642, applied this same technique to identify the portion of the human genome which is bound by the GLI gene product produced as a recombinant fusion protein. The GLI gene is amplified in a subset of human tumors. Ellington, A.D., et al., Nature (1990) 346: 818-822, describe the production of a large number of random sequence RNA molecules and identification of those which bind specifically to immobilized target molecules, in the case of this paper, to specific dyes such as Cibacron blue. Randomly synthesized DNA yielding approximately 10 individual sequences was amplified by
PCR and transcribed into RNA. It was thought that the complexity of the pool was reduced in the amplification/transcription steps to approximately 10 different sequences. The pool was then applied to an 5 affinity column containing the dye and the bound sequences subsequently eluted, treated with reverse transcriptase and amplified by PCR. The results showed that about one in 10 random sequence RNA molecules folds in such a way as to bind specifically to the
10 ligand.
Thiesen, H.-J., and Bach, C, Nucleic Acids Res. (1990) 11:3203-3208, describe what they call a target detection assay (TDA) to determine double-stranded DNA binding sites for putative DNA binding proteins. In
15 their approach, a purified functionally active DNA binding protein and a pool of random double-stranded oligonucleotides which contain PCR primer sites at each end were incubated with the protein. The resulting DNA complexes with the protein (in their case, the SP-1
20 regulatory protein) were separated from the unbound oligomers in the random mixture by band-shift electrophoresis and the SP-1 bound oligonucleotides were rescued by PCR and cloned, and then sequenced.
None of the cited references describe the use
25 of single-stranded DNA as an appropriate material for generating aptamers. The use of DNA aptamers has several advantages over RNA including increased nuclease stability (Shaw, J.P. et al., Nuc Acid Res (1991) 19:747- 750) , in particular plasma nuclease stability, and ease
≠ 30 of amplificatio: by PCR or other methods. RNA generally is converted to DNA prior to amplification using reverse transcriptase, a process that is not equally efficient with all sequences, resulting in loss of some aptamers from a selected pool.
35
Finally, none of the above references describes (i) the identification of oligonucleotides which specifically bind to thrombin, which does not normally bind to DNA; (ii) interference with the normal biological function of target molecules such as thrombin due to binding; (iii) the use of linkages other than the standard phosphodiester linkages in the backbone of the oligonucleotide, (iv) the use of base analogs in the oligonucleotide, (v) target-specific binding of short aptamer sequences and aptamer analog sequences derived from a larger full-length parent aptamer molecule, (vi) in vivo therapeutic efficacy of an aptamer or (vii) in vivo therapeutic efficacy of an aptamer analog. Thrombin. Acute vascular diseases are associated with partial or total occlusion of a blood vessel by blood clots, which contain platelets and fibrin. These diseases include serious health risks such as myocardial infarction, deep vein thrombosis, pulmonary embolism, peripheral arterial occlusion and the like. Treatment or prophylaxis of thrombotic diseases is based on either inhibition of clotting or acceleration of thrombolysis. Both approaches to treatment of thrombotic disease have been described using agents such as heparin or hirudin to inhibit thrombin and streptokinase or tissue plasminogen activator to accelerate thrombolysis. However, a need remains for improved therapeutic agents that inhibit tne activities of thrombin in clot formation, platelet aggregation or activation and other thrombin-mediated processes. Thrombin is a multifunctional enzyme that (i) converts fibrinogen to fibrin by enzymatic cleavage; (ii) has mitogenic ef ects on lymphocytes and vascular smooth muscle cells; (iii) stimulates platelet aggregation and activation; (iv) is chemotactic for monocytes; (v) stimulates vascular endothelial cell mediated production
of prostacyclin, platelet-activating factor and other factors; (vi) induces neutrophil adherence to vessel walls; (vii) stimulates vascular endothelial cell adhesion phenotype; and (viii) generates activated protein C by cleavage of protein C.
Mitogenic activity of thrombin is exerted through binding to thrombin receptors (Coughlin, S.R., et al, J. Clin. Invest.. (1992) 81:351-355). Platelet aggregation, which plays a major role in arterial thrombosis is largely dependent on the function of thrombin (Hanson, S.R., et al, Proc. Natl. Acad. Sci. USA. (1988) £5_:3184-3188) . Platelets carry functional thrombin receptors. Inflammatory responses can also be mediated by thrombin through stimulation of platelet activating factor (PAF) (Prescott, S., et al, Proc. Natl. Acad. Sci. USA. (1984) £1:3534-3538. PAF promotes adhesion of neutrophils to endothelial matrix, leading to degranulation of the neutrophils and an associated inflammatory response.
Disclosure of the Invention
The identification of oligonucleotides that specifically bind to thrombin, which does not normally bind to RNA or DNA, has now been demonstrated. The thrombin aptamers bind to thrombin and inhibit both its catalytic activity in converting fibrinogen to fibrin and its platelet aggregating activity. The aptamers are potent inhibitors of thrombin function and represent a new cla* of pharmaceutical agents for modulation of the activity of this protease. The molecules of this invention may be utilized in compositions and methods for inhibiting any thrombin-mediated or thrombin-associated process or function. Pharmaceutical compositions containing these molecules, as well as methods of treatment or prophylaxis of vascular diseases,
inflammatory responses, cancer-related hypercoagulable states, sepsis and neural vasooclusive diseases using these compositions are also part of the present invention. These molecules can also be utilized in compositions and methods for in vitro or in vivo imaging, diagnosis, for storing and treating extracorporeal blood and for coating implant devices.
These molecules can be synthesized chemically or enzymatically as described below, and can be prepared in commercial quantities. The aptamers of the present invention are composed of DNA and chemically related molecules. DNA is a class of molecule ordinarily found in animals and it is expected that the immunogenicity of thrombin aptamers will be nonexistent or very low. Immune reactions against nucleic acids are known to be rare and, when observed, are associated with autoimmune disorders. Because of their compatibility with biological systems, the molecules of the invention are suitable in the treatment of both acute and nonacute vascular conditions.
In one aspect, the invention is directed to a method to determine an aptamer which binds specifically to thrombin, which method comprises providing a mixture containing oligomers optionally having portions which form a random set of sequences and portions which permit amplification of the oligomers, incubating the oligomer mixture with thrombin coupled to a support to form complexes between thrombin and the oligomers bound specifically thereto, removing the unbound members of the oligonucleotide mixture from the support environment, recovering the complexεd oligonucleotide(s) from the support, amplifying the recovered oligonucleotides, and sequencing the recovered and amplified oligonucleotide(s; which had been complexed with thrombin. In a preferred embodiment, the starting mixture of oligonucleotides
having random sequences may also contains a consensus sequence known to bind to thrombin.
In yet another aspect, this invention is directed to single-stranded deoxyribonucleotides that bind specifically to thrombin. It has been heretofore thought that the three-dimensional structure of double- stranded DNA limited the structural diversity of the molecule. The inventors herein are unaware of any prior demonstration of structural diversity for single- or double-stranded DNA sufficient to provide the range of conformations necessary to provide aptamers to biomolecules. For example, known RNA structures, such as pseudoknots, have not been described for single-stranded DNA. In other aspects, the invention is directed to oligonucleotides which contain sequences identified by the above methods, and to oligonucleotide sequences which bind specifically to thrombin. In still another aspect, the invention is directed to complexes comprising the thrombin target substance and specifically bound oligomer.
In still other aspects, the invention is directed to oligomers which contain sequences that bind specifically to thrombin target substances and inhibit its normal biological function, and to the use of these oligomers in therapy, diagnostics, and purification procedures.
In yet a further aspect, this invention is directed to oligomers which contain sequences that bind specifically to thrombin and inhibits its normal biological function, and which also contain one or more modified bases, sugars, or sugar linkages, and to the use of these oligomers in therapy, diagnostics, and purification procedures.
Brief Description of the Figures
Figure 1 is a chart depicting thrombin aptamer consensus-related sequences.
Figure 2 is a plot of in vivo thrombin 5 inhibition obtained from primates using a 15-mer aptamer.
Modes of Carrying Out the Invention
The practice of the present invention encom¬ passes conventional techniques of chemistry, molecular 0 biology, biochemistry, protein chemistry, and recombinant DNA technology, which are within the skill of the art. Such techniques are explained fully in the literature. See, e.g., Oligonucleotide Synthesis (M.J. Gait ed. 1984); Nucleic Acid Hybridization (B.D. Hames & S.J. 5 Higgins, eds., 1984); Sambrook, Fritsch & Maniatis,
Molecular Cloning; A Laboratory Manual. Second Edition (1989); PCR Technology (H.A. Erlich ed. , Stockton Press); R.K. Scope, Protein Purification Principles and Practice (Springer-Verlag) ; and the series Methods in Enzymology 0 (S. Colowick and N. Kaplan eds.. Academic Press, Inc.). All patents, patent applications and publications mentioned herein, whether supra or infra, are hereby incorporated by reference in their entirety. The invention is directed to a method which 5 permits the recovery and deduction or identification of aptamers which bind specifically to thrombin and compositions that result from the use of the method.
For example, these aptamers can be used as a separation tool for retrieving or detecting thrombin. In G these methods, the aptamers function much like monoclonal antibodies in their specificity and usage. By coupling the aptamers containing the specifically binding sequences to a solid support, thrombin can be recovered in useful quantities. In addition, these aptamers can be t
used in diagnosis by employing them in specific binding assays.
For application in such various uses, the aptamers of the invention may be coupled to auxiliary substances that enhance or complement the function of the aptamer. Such auxiliary substances include, for example, labels such as radioisotopes, fluorescent labels, enzyme labels and the like; specific binding reagents such as antibodies, additional aptamer sequence, cell surface receptor ligands, receptors per se and the like; toxins such as diphtheria toxin, tetanus toxin or ricin; drugs such as antiinflammatory, antibiotic, or metabolic regulator pharmaceuticals, solid supports such as chromatographic or electrophoretic supports, and the like. Suitable techniques for coupling of aptamers to desired auxiliary substances are generally known for a variety of such auxiliary substances, and the specific nature of the coupling procedure will depend on the nature of the auxiliary substance chosen. Coupling may be direct covalent coupling or may involve the use of synthetic linkers such as those marketed by Pierce Chemical Co., Rockford, IL.
As used herein, "specifically binding oligonucleotides" or "aptamers" refers to oligonucleotides having specific binding regions which are capable of forming complexes with thrombin in an environment wherein other substances in the same environment are not complexed to the oligonucleotide. The specificity of the binding is defined in terms of the comparative dissociation constants of the. aptamer for thrombin as compared to the dissociation constant with respect to the aptamer and other materials in the environment or unrelated molecules in general. Typically, the Kd for the aptamer with respect to thrombin will be 2-fold, preferably 5-fold, more
preferably 10-fold less than Kd with respect to thrombin and the unrelated material or accompanying material in the environment. Even more preferably the Kd will be 50- fold less, more preferably 100-fold less, and more preferably 200-fold less.
The binding affinity of the aptamers herein with respect to thrombin is defined in terms of Kd. The value of this dissociation constant can be determined directly by well-known methods, and can be computed even for complex mixtures by methods such as those, for example, set forth in Caceci, M. , et al., Byte (1984) 9_:340-362. It has been observed, however, that for some small oligonucleotides, direct determination of Kd is difficult, and can lead to misleadingly high results. Under these circumstances, a competitive binding assay for thrombin may be conducted with respect to substances known to bind thrombin. The value of the concentration at which 50% inhibition occurs (Ki) is, under ideal conditions, equivalent to Kd. However, in no event can Ki be less than Kd. Thus, determination of Ki, in the alternative, sets a maximal value for the value of Kd. Under those circumstances where technical difficulties preclude accurate measurement of Kd, measurement of Ki can conveniently be substituted to provide an upper limit for Kd.
As specificity is defined, in terms of Kd as set forth above, excluded from the categories of unrelated materials and materials accompanying thrombin in its environment are those materials which are sufficiently related to thrombin to be immunologically crossreactive therewith, and materials which natively bind oligonucleotides of particular sequences such as nucleases, restriction enzymes, and the like. By "immunologically crossreactive" is meant that antibodies raised with respect to thrombin crossreact under standard
assay conditions with the candidate material. Generally, for antibodies to crossreact in standard assays, the binding affinities of the antibodies for crossreactive materials as compared to thrombin should be in the range of 5-fold to 100-fold, generally about 10-fold.
Thus, aptamers which contain specific binding regions are specific with respect to unrelated materials and with respect to materials which do not normally bind such oligonucleotides such as nucleases and restriction enzymes. In general, a minimum of approximately 6 nucleotides, preferably 10, and more preferably 14 or 15 nucleotides, are necessary to effect specific binding. Aptamers of sequences as short as 6 bases have been shown to specifically bind and inhibit thrombin. The only apparent limitations on the binding specificity of the thrombin/oligonucleotide couples of the invention concern sufficient sequence to be distinctive in the binding oligonucleotide and sufficient binding capacity of thrombin to obtain the necessary interaction. Oligonucleotides of sequences shorter than 10, e.g., 6 mers, are feasible if the approp* ate interaction can be obtained in the context of the environment in which the thrombin is placed. Thus, if there are few interferences by other materials, less specificity and less strength of binding may be required.
As used herein, "aptamer" refers in general to either an oligonucleotide of a single defined sequence or a mixture of said oligonucleotides, wherein the mixture retains the properties of binding specifically to thrombin. Thus, as used herein "aptamer" denotes both singular and plural sequences of oligonucleotides, as defined herein.
Structurally, the aptamers of the invention are specifically binding oligonucleotides, wherein "oligonucleotide" is as defined herein. As set forth
herein, oligonucleotides include not only those with conventional bases, sugar residues and internucleotide linkages, but also those which contain modifications of any or all of these three moieties. "Single-stranded" oligonucleotides, as the term is used herein, refers to those oligonucleotides which contain a single covalently linked series of nucleotide residues.
"Oligomers" or "oligonucleotides" include RNA or DNA sequences of more than one nucleotide in either single chain or duplex form and specifically includes short sequences such as di ers and trimers, in either single chain or duplex form, which may be intermediates in the production of the specifically binding oligonucleotides.
"Oligonucleotide" or "oligomer" is generic to polydeoxyribonucleotides (containing 2'-deoxy-D-ribose or modified forms thereof), i.e., DNA, to polyribonucleo- tides (containing D-ribose or modified forms thereof) , i.e., RNA, and to any other type of polynucleotide which is an N-glycoside or C-glycoside of a purine or pyrimidine base, or modified purine or pyrimidine base. The oligomers of the invention may be formed using conventional phosphodiester-linked nucleotides and synthesized using standard solid phase (or solution phase) oligonucleotide synthesis techniques, which are now commercially available. However, the oligomers of the invention may also contain one or more "substitute" linkages as is generally understood in the art. Some of these substitute linkages are non-polar and contribute to the desired ability of the oligomer to diffuse across membranes. These "substitute" linkages are defined herein as conventional alternative linkages such as phosphorothioate or phosphoramidate, are synthesized as described in the generally available literature.
Alternative linking groups include, but are not limited to embodiments wherein a moiety of the formula P(0)S, ("thioate"), P(S)S ("dithioate") , P(0)NR'2, P(0)R', P(0)0R , CO, or CONR',, wherein R' is H (or a salt) or alkyl (1-12C) and RD is alkyl (1-9C) is joined to adjacent nucleotides through -0- or -S-. Dithioate linkages are disclosed and claimed in commonly owned U.S. application no. 248,517. Substitute linkages that may be used in the oligomers disclosed herein also include nonphosphorous-based internucleotide linkages such as the 3' -thioformacetal (-S-CH2-0-), formacetal (-0-CH2-0-) and 3' -amine (-NH-CH2-CH2-) internucleotide linkages disclosed and claimed in commonly owned pending U.S. patent application serial nos. 690,786 and 763,130, both incorporated herein by reference. One or more substitute linkages may be utilized in the oligomers in order to further facilitate binding with complementary target nucleic acid sequences or to increase the stability of the oligomers toward nucleases, as well as to confer permeation ability. (Not all such linkages in the same oligomer need be identical.)
The term "nucleoside" or "nucleotide" is similarly generic to ribonucleosides or ribonucleotides, deoxyribonucleosides or deoxyribonucleotides, or to any other nucleoside which is an N-glycoside or C-glycoside of a purine or pyrimidine base, or modified purine or pyrimidine base. Thus, the stereochemistry of the sugar carbons may be other than that of D-ribose in one or more residues. Also included are analogs where the ribose or deoxyribose moiety is replaced by an alternate structure such as the 6-membered morpholino ring described in U.S. patent number 5,034,506 or where an acyclic structure serves as a scaffold that positions the base analogs described herein in a manner that permits efficient binding to target nucleic acid sequences or other
targets. Elements ordinarily found in oligomers, such as the furanose ring or the phosphodiester linkage may be replaced with any suitable functionally equivalent element. As the a anomer binds to targets in a manner similar to that for the β anomers, one or more nucleotides may contain this linkage or a domain thereof. (Praseuth, D. , et al., Proc Natl Acad Sci (USA) (1988) £5:1349-1353) . Modifications in the sugar moiety, for example, wherein one or more of the hydroxyl groups are replaced with halogen, aliphatic groups, or functionalized as ethers, amines, and the like, are also included.
"Nucleoside" and "nucleotide" include those moieties which contain not only the natively found purine and pyrimidine bases A, T, C, G and U, but also modified or analogous forms thereof. Modifications include alkylated purines or pyrimidines, acylated purines or pyrimidines, or other heterocycles. Such "analogous purines" and "analogous pyrimidines" are those generally known in the art, many of which are used as chemotherapeutic agents. An exemplary but not exhaustive list includes pseudoisocytosme, N 4,N4-ethanocytosine, 8- hydroxy-N -methyladenine, 4-acetylcytosine, *5- (carboxyhydroxylmethyl) uracil, 5-fluorouracil, 5-bromouracil, 5-carboxymethylaminomethyl-2-thiouracil, 5-carboxymethylaminomethyl uracil, dihydrouracil, inosine, N -iscpentenyl-adenine, 1-methyladenine, 1-methylpseudouracil, 1-methylguanine, 1-methylinosine, 2,2-dimethylguanine, 2-methyladenine, 2-methylguanine, 3- methylcytosine, 5-methylcytosine, N -methyladenine, 7- methylguanine, 5-methylaminomethyl uracil, 5-methoxy aminomethyl-2-thiouracil, beta-D-mannosylqueosine, 5'- methoxycarbonylmethyluracil, 5-methoxyuracil, 2- methylthio-N -isopentenyladenine, uracil-5-oxyacetic ac methyl ester, pseudouracil, 2-thiocytosine, 5-methyl-2-
thiouracil, 2-thiouracil, 4-thiouracil, 5-methyluracil, N-uracil-5-oxyacetic acid methylester, uracil-5-oxyacetic acid, queosine, 2-thiocytosine, 5-propyluracil, 5-propylcytosine, 5-ethyluracil, 5-ethylcytosine, 5-butyluracil, 5-butylcytosine, 5-pentyluracil, 5-pentylcytosine, and 2,6-diaminopurine.
In addition to the modified bases above, nucleotide residues which are abasic, i.e., devoid of a purine or a pyrimidine base may also be included in the aptamers of the invention and in the methods for their obtention.
The sugar residues in the oligonucleotides of the invention may also be other than conventional ribose and deoxyribose residues. In particular, substitution at the 2' -position of the furanose residue is particularly important.
Aptamer oligonucleotides may contain analogous forms of ribose or deoxyribose sugars that are generally known in the art. An exemplary, but not exhaustive list includes 2' substituted sugars such as 2' -O-methyl-, 2'- 0-alkyl, 2'-0-allyl, 2'-S-alkyl, 2'-S-allyl, 2'-fluoro-, 2' -halo, or 2' -azido-ribose, carbocyclic sugar analogs, Qf-anomeric sugars, epimeric sugars such as arabinose, xyloses or lyxoses, pyranose sugars, furanose sugars, sedoheptuloses, acyclic analogs and abasic nucleoside analogs such as methyl riboside, ethyl riboside or propyl riboside.
.Although the conventional sugars and bases will be used in applying the method of the invention, substi- tution of analogous forms of sugars, purines and pyrimidines can be advantageous in designing the final product. Additional techniques, such as methods of synthesis of 2'-modified sugars or carbocyclic sugar analogs, are described in Sproat, B.S. et al., Nuc Acid Res (1991) l£:733-738; Cotten, M. et al. , Nuc Acid Res
(1991) 19_:2629-2635; Hobbs, J. et al., Biochemistry (1973) 12:5138-5145; and Perbost, M. et al., Biochem Biophys Res Comm (1989) 165:742-747 (carbocyclics) .
Methods to Prepare the Invention Aptamers
In general, the method for preparing the aptamers of the invention involves incubating thrombin with a mixture of oligonucleotides under conditions wherein some but not all of the members of the oligonucleotide mixture form complexes with the thrombin. The resulting complexes are then separated from the uncomplexed members of the oligonucleotide mixture and the complexed members which constitute an aptamer (at this stage the aptamer generally being a population of a multiplicity of oligonucleotide sequences) is recovered from the complex and amplified. The resulting aptamer (mixture) may then be substituted for the starting mixture in repeated iterations of this series of steps. When satisfactory specificity is obtained, the aptamer may be used as obtained or may be sequenced and synthetic forms of the aptamer prepared. In this most generalized form of the method, the oligonucleotides used as members of the starting mixture may be single-stranded or double- stranded DNA or RNA, or modified forms thereof. However, single-stranded DNA is preferred. The use of DNA eliminates the need for conversion of RNA aptamers to DNA by reverse transcriptase prior to PCR amplification. Furthermore, DNA is less susceptible to nuclease degradation than RNA. The oligonucleotides that bind to thrombin are separated from the rest of the mixture and recovered and amplified. Amplification may be conducted before or after separation from thrombin. The oligonucleotides are conveniently amplified by PCR to give a pool of DNA sequences. The PCR method is well known in the art and
described in, e.g., U.S. Patent Nos. 4,683,195 and 4,683,202 and Saiki, R.K., et al. , Science (1988) 3£:487-491, and European patent applications 86302298.4, 86302299.2 and 87300203.4, as well as Methods in Enzvmologv (1987) 15_5_:335-350. If RNA is initially used, the amplified DNA sequences are transcribed into RNA. The recovered DNA or RNA, in the original single-stranded or duplex form, is then used in another round of selection and amplification. After three to six rounds of selection/amplification, oligomers that bind with an affinity in the mM to μM range can be obtained and affinities below the μM range are possible. PCR may also be performed in the presence of thrombin.
Other methods of amplification may be employed including standard cloning, ligase chain reaction, etc. (See e.g., Chu, et al., U.S. Patent No. 4,957,858). For example, to practice this invention using cloning, once the aptamer has been identified, linkers may be attached to each side to facilitate cloning into standard vectors. Aptamers, either in single or double stranded form, may be cloned and recovered thereby providing an alternative amplification method.
Amplified sequences can be applied to sequencing gels after any round to determine the nature of the aptamers being selected by thrombin. The entire process then may be repeated using the recovered and amplified duplex if sufficient resolution is not obtained.
Amplified sequences can be cloned and individual oligonucleotides then sequenced. The entire process can then be repeated using the recovered and amplified oligomers as needed. Once an aptamer that binds specifically to thrombin has been selected, it may be recovered as DNA or RNA in single-stranded or duplex form using conventional techniques.
Similarly, a selected aptamer may be sequenced and resynthesized using one or more modified bases, sugars and linkages using conventional techniques. The specifically binding oligonucleotides need to contain the sequence-conferring specificity, but may be extended with flanking regions and otherwise derivatized.
The starting mixture of oligonucleotide may be of undetermined sequence or may preferably contain a randomized portion, generally including from about 3 to about 400 nucleotides, more preferably 10 to 100 nucleotides. The randomization may be complete, or there may be a preponderance of certain sequences in the mixture, or a preponderance of certain residues at particular positions. Although, as described hereinbelow, it is not essential, the randomized sequence is preferably flanked by primer sequences which permit the application of the polymerase chain reaction directly to the recovered oligonucleotide from the complex. The flanking sequences may also contain other convenient features, such as restriction sites which permit the cloning of the amplified sequence. These primer hybridization regions generally contain 10 to 30, more preferably 15 to 25, and most preferably 18 to 20, bases of known sequence. The oligonucleotides of the starting mixture may be conventional oligonucleotides, most preferably single-stranded DNA, or may be modified forms of these conventional oligomers as described hereinabove. For oligonucleotides containing conventional phosphodiester linkages or closely related forms thereof, standard oligonucleotide synthesis techniques may be employed. Such techniques are well known in the art, such methods being described, for example, in Froehler, B., et al., Nucleic Acids Research (1986) 14:5399-5467; Nucleic Acids Research (1988) 16:4831-4839; Nucleosides and Nucleotides
(1987) .6:287-291; Froehler, B., Tet Lett (1986) 22:5575- 5578. Oligonucleotides may also be synthesized using solution phase methods such as triester synthesis, known in the art. The nature of the mixture is determined by the manner of the conduct of synthesis. Randomization can be achieved, if desired, by supplying mixtures of nucleotides for the positions at which randomization is desired. Any proportion of nucleotides and any desired number of such nucleotides can be supplied at any particular step. Thus, any degree of randomization may be employed. Some positions may be randomized by mixtures of only two or three bases rather than the conventional four. Randomized positions may alternate with those which have been specified. It may be helpful if some portions of the candidate randomized sequence are in fact known.
In one embodiment of the method of the invention, the starting mixture of oligonucleotides subjected to the invention method will have a binding affinity for thrombin characterized by a Kd of 1 μM or greater. Binding affinities of the original mixture for thrombin may range from about 100 μM to 10 μM to 1 μM but, of course, the smaller the value of the dissociation constant, the more initial affinity there is in the starting material for thrombin. This may or may not be advantageous as specificity may be sacrificed by starting the procedure with materials with high binding affinity. By application of the method of the invention as described herein, improvements in the binding affinity over one or several iterations of the above steps of at least a factor of 50, preferably of a factor of 100, and more preferably of a factor of 200 may be achieved. As defined herein, a ratio of binding affinity reflects the ratio of Kds of the comparative complexes. Even more preferred in the conduct of the method of the invention
is the achievement of an enhancement of an affinity of a factor of 500 or more.
Thus, the method of the invention can be conducted to obtain the invention aptamers wherein the 5 aptamers are characterized by consisting of single- stranded DNA, by having a binding affinity for thrombin represented by a Kd of 100 x 10 or less, by having a specificity representing by a factor of at least 2, and preferably 5, more preferably 10 with respect to 0 unrelated molecules, by having a binding region of less than 15 nucleotide residues or a total size of less than 16 nucleotide residues, or by binding to thrombin.
The invention processes are also characterized by accommodating starting mixtures of oligonucleotides 5 having a binding affinity for thrombin characterized by a Kd of l μM or more by an enhancement of binding affinity of 50 or more, and by being conducted under physiological conditions.
.As used herein, physiological conditions means 0 the salt concentration and ionic strength in an aqueous solution which characterize fluids found in human metabolism commonly referred to as physiological buffer or physiological saline. In general, these are represented by an intracellular pH of 7.1 and salt 5 concentrations Na+:5-15 mM, K+:140 mM, Mg+2:0.3 mM,
Ca+ :10"4 mM, Cl":5-13 mM, and an extracellular pH of 7.4 and salt concentrations Na+:145 mM, K+:3 mM, Mg :l-2 mM, Ca+2:l-2 mM; Cl":110 mM.
The use of physiological conditions in the C aptamer selection method is extremely important, particularly with respect to those aptamers that may be intended for therapeutic use. .As is understood in the art, the concentration of various ions, in particular, the ionic strength, and the pH value impact on the value
of the dissociation constant of the thrombin/aptamer complex.
Use of Modified Nucleotides and Oligonucleotides In one embodiment of the invention method, the initial mixture of candidate oligonucleotides will include oligc. ars which contain at least one modified nucleotide residue or linking group.
If certain specific modifications are included in the amplification process as well, advantage can be taken of additional properties of any modified nucleotides, such as the presence of specific affinity agents in the purification of the desired materials. In order for the modified oligomer to yield useful results, the modification must result in a residue which is "read" in a known way by the polymerizing enzyme used in the amplification procedure. It is not necessary that the modified residue be incorporated into the oligomers in the amplification process, as long it is possible to discern from the nucleotide incorporated at the corresponding position the nature of the modification contained in the candidate, and provided only one round of complexation/amplification is needed. However, many of the modified residues of the invention are also susceptible to enzymatic incorporation into oligonucleotides by the commonly used polymerase enzymes and the resulting oligomers will then directly read on the nature of the candidate actually contained in the initial complex. It should be noted that if more than one round of complexation is needed, the amplified sequence must include the modified residue, unless the entire pool is sequenced and resynthesized to include the modified residue.
Certain modifications can be made to the base residues in a oligonucleotide sequence without impairing
the function of polymerizing enzymes to recognize the modified base in the template or to incorporate the modified residue. These modifications include alkylation of the 5-position of uridine, deoxyuridine, cytidine and deoxycytidine; the N -position of cytidine and deoxycytidine; the N -position of adenine and deoxyadenine; the 7-position of 7-deazaguanine, 7- deazadeoxyguanine, 7-deazaadenine and 7-deazadeoxy- adenine. As long as the nature of the recognition is known, the modified base may be included in the oligomeric mixtures useful in the method of the invention.
The nature of the sugar moiety may also be modified without affecting the capacity of the sequence to be usable as a specific template in the synthesis of new DNA or RNA.
The efficacy of the process of selection and amplification depends on the ability of the PCR reaction faithfully to reproduce the sequence actually complexed to thrombin. Thus, if the oligonucleotide contains modified forms of cytosine (C*) , the PCR reaction must recognize this as a modified cytosine and yield an oligomer in the cloned and sequenced product which reflect this characterization. If the modified form of cytosine (C*) is included in the PCR reaction as dC*TP, the resulting mixture will contain C* at positions represented by this residue in the original member of the candidate mixture. (It is seen that the PCR reaction cannot distinguish between various locations of C* in the original candidate; all C residue locations will appear as C*.) Conversely, dCTP could be used in the PCR reaction and it would be understood that one or more of the positions now occupied by C was occupied in the
* original candidate mixture by C , provided only one round of complexation/amplification is needed. If the
amplified mixture is used in a second round, this new mixture must contain the modification.
Of course, if the selected aptamer is sequenced and resynthesized, modified oligonucleotides and linking groups may arbitrarily be used in the synthesized form of the aptamer.
Inclusion of modified oligonucleotides in the methods and aptamers of the invention provides a tool for expansion of the repertoire of candidates to include large numbers of additional oligonucleotide sequences. Such expansion of the candidate pool may be especially important as the demonstration of binding to proteins, for example, in the prior art is limited to those proteins known to have the capability to bind DNA. Modifications of the oligonucleotide may be necessary to include all desired sequences among those for which specific binding can be achieved.
Thus, one preferred method comprises incubating thrombin with a mixture of oligonucleotides, wherein these oligonucleotides contain at least one modified nucleotide residue or linkage, under conditions wherein complexation occurs with some but not all members of the mixture; separating the complexed from uncomplexed oligonucleotides, recovering and amplifying the complexed oligonucleotides and optionally determining the sequence of the recovered nucleotides. In an additional preferred embodiment, amplification is also conducted in the presence of modified nucleotides.
A Subtraction Method for Aptamer Preparation
It is often advantageous in enhancing the specificity of the aptamer obtained to remove members of the starting oligonucleotide mixture which bind to a second substance from which thrombin is to be distinguished. In such subtraction methods, at least two
rounds of selection and amplification will be conducted. In a positive/negative selection approach, thrombin will be incubated with the starting mixture of oligonucleotides and, as usual, the complexes form separated from uncomplexed oligonucleotides. The complex oligonucleotides, which are now an aptamer, are recovered and amplified from the complex. The recovered aptamer is then mixed with the second undesired substance from which thrombin is to be distinguished under conditions wherein members of the aptamer population which bind to said second substance can be complexed. This complex is then separated from the remaining oligonucleotides of the aptamer. The resulting second uncomplexed aptamer population is then recovered and amplified. The second aptamer population is highly specific for thrombin as compared to the second substance.
In an alternative approach, the negative selection step may be conducted first, thus mixing the original oligonucleotide mixture with the undesired substance to complex away the members of the oligonucleotide mixture which bind to the second substance; the uncomplexed oligonucleotides are then recovered and amplified and incubated with thrombin under conditions wherein those members of the oligonucleotide mixture which bind thrombin are complexed. The resulting complexes then removed from the uncomplexed oligonucleotides and the bound aptamer population is recovered and amplified as usual.
Modified Method Wherein Thrombin/Aptamer Complexes are Separated from Solid Support
As set forth hereinabove, the original oligonucleotide mixture can be synthesized according to the desired contents of the mixture and can be separated by adding the oligonucleotide mixture to a column
containing covalently attached thrombin (see, Ellington, A.D., et al., Nature (1990) 346:818-822) or to thrombin in solution (see Blackwell et al., Science (1990) 25_0:1104-1110; Blackwell et al., Science (1990) 250:1149- 1151; or to thrombin bound to a filter (see Tuerk, C. , and Gold, L. , Science (1990) 249:505-510) . Complexes between the aptamer and thrombin are separated from uncomplexed aptamers using any suitable technique, depending on the method used for complexation. For example, if columns are used, non-binding species are simply washed from the column using an appropriate buffer. Specifically b~und material can then be eluted.
If binding occurs in solution, the complexes can be separated from the uncomplexed oligonucleotides using, for example, the mobility shift in electrophoresis technique (EMSA) , described in Davis, R.L., et al., Cell (1990) 6J1:733. In this method, aptamer-thrombin complexes are run on a gel and aptamers removed from the region of the gel where thrombin runs. Unbound oligomers migrate outside these regions and are separated away. Finally, if complexes are formed on filters, unbound aptamers are eluted using standard techniques and the desired aptamer recovered from the filters.
In a preferred method, separation of the complexes involves detachment of thrombin-aptamer complexes from column matrices as follows.
A column or other support matrix having covalently or noncovalently coupled thrombin is synthesized. Any standard coupling reagent or procedure may be utilized, depending on the nature of the support. For example, covalent binding may include the formation of disulfide, ether, ester or amide linkages. The length of the linkers used may be varied by conventional means. Noncovalent linkages include antibody-antigen interactions, protein-sugar interactions, as between, for
example, a lectin column and a naturally-occurring oligosaccharide unit on a peptide.
Lectin columns are particularly suited for selecting thrombin aptamers. Lectins are proteins or glycoproteins that can bind to complex carbohydrates or oligosaccharide units on glycoproteins, and are well- described in The Lectins (I.E. Liener et al., eds.. Academic Press 1986) . Lectins are isolated from a wide variety of natural sources, including peas, beans, lentils, pokeweed and snails. Concanavalin A is a particularly useful lectin.
Other linking chemistries are also available. For example, disulfide-derivatized biotin (Pierce) may be linked to thrombin by coupling through an amine or other functional group. The resulting thrombin-S-S-biotin complex could then be used in combination with avidin- derivatized support. Oligonucleotide-thrombin complexes could then be recovered by disulfide bond cleavage. Linking chemistries will be selected on the basis of (i) conditions or reagents necessary for maintaining the structure or activity of thrombin.
The oligomer mixture is added to and incubated with the support to permit oligonucleotide-thrombin complexation. Complexes between the oligonucleotides and thrombin are separated from uncomplexed oligonucleotides by removing unbound oligomers from the support environment. For example, if columns are used, nonbinding species are simply washed from the column using an appropriate buffer. Following removal of unbound oligomers, the thrombin is uncoupled from the support. The uncoupling procedure depends on the nature of the coupling, as described above. Thrombin bound through disulfide linkages, for example, may be removed by adding a sulfhydryl reagent such as dithiothreitol or β-
mercaptoethanol. Thrombin bound to lectin supports may be removed by adding a complementary monosaccharide (e.g., α-methyl-mannoside, N-acetyl glucosamine, glucose, N-acetyl galactosamine, galactose or other saccharides for concanavalin A) . Oligonucleotides specifically bound to thrombin can then be recovered by standard denaturation techniques such as phenol extraction.
The method of elution of thrombin- oligonucleotide complex from a support has superior unexpected properties when compared with standard oligonucleotide elution techniques. This invention is not dependent on the mechanism by which these superior properties occur. However, without wishing to be limited by any one mechanism, the following explanation is offered as to how more efficient elution is obtained. Certain support effects result from the binding of oligonucleotides to the support, or the support in conjunction with oligonucleotide or thrombin. Removing oligonucleotide-thrombin complexes enables the recovery of oligonucleotides specific to thrombin only, while eliminating oligonucleotides binding to the support, or the support in conjunction with oligonucleotide or thrombin. At each cycle of selection, this method may give up to 1,000-fold enrichment for specifically binding species. Selection with thrombin remaining bound to support gives less enrichment per cycle, making it necessary to go through many more cycles in order to get a good aptamer population.
Aptamer Pools of Varying Length
Aptamers can also be selected in the above methods using a pool of oligonucleotides that vary in length as the starting material. Thus, several pools of oligonucleotides having random sequences are synthesized that vary in length from e.g. 50 to 60 bases for each
pool and containing the same flanking primer-binding sequences. Equal molar amounts of each pool are mixed and the variable-length pool is then used to select for aptamers that bind to thrombin, as described above. This protocol selects for the optimal species for thrombin binding from the starting pool and does not limit aptamers to those of a given length.
Alternatively, several pools of mixed length aptamers can be used in parallel in separate selections and then combined and further selected to obtain the optimal binders from the size range initially used. For example, three pools, A, B and C, can be used. Pool A can consist of oligonucleotides having random sequences that vary in length from e.g. 30 to 40 bases; pool B can have sequences varying in length from e.g. 40 to 50 bases; and pool C can have sequences varying in length from 50 to 60 bases. It is to be understood that the lengths described above are for illustrative purposes only. After selection to obtain binders from A, B, and C, all aptamers are mixed together. A number of rounds of selection are done as described above to obtain the best binders from the initial species selected in the 30- to 60-base range. Note that with this technique, not all possible species in some of the pools are used for selection. If the number of sites available for binding are increased, i.e., if a column is used and the size of the column increased, more species can be included for selection. Furthermore, this method allows for the selection of oligomers from the initial starting pool that are of optimal length for binding thrombin.
Derivatization
Aptamers containing the specific binding sequences discerned through the method of the invention can also be derivatized in various ways. For example, if
the aptamer is to be used for separation of thrombin, conventionally the oligonucleotide will be derivatized to a solid support to permit chromatographic separation. If the oligonucleotide is to be used for attaching a detectable moiety to thrombin, the oligonucleotide will be derivatized to include a radionuclide, a fluorescent molecule, a chromophore or the like. If the oligonucleo¬ tide is to be used in specific binding assays, coupling to solid support or detectable label, and the like are also desirable. If it is to be used in therapy, the oligonucleotide may be derivatized to include ligands which permit easier transit of cellular barriers, toxic moieties which aid in the therapeutic effect, or enzymatic activities which perform desired functions at the thrombin site. The aptamer may also be included in a suitable expression system to provide for in situ generation of the desired sequence.
Consensus Sequences When a number of individual, distinct aptamer sequences for thrombin have been obtained and sequenced as described above, the sequences may be examined for "consensus sequences." As used herein, "consensus sequence" refers to a nucleotide sequence or region (which may or may not be made up of contiguous nucleotides) , which is found in one or more regions of at least two aptamers, the presence of which may be correlated with aptamer-to-thrombin-binding or with aptamer structure. A consensus sequence may be as short as three nucleotides long. It also may be made up of one or more noncontiguous sequences with nucleotide sequences or polymers of hundreds of bases long interspersed between the consensus sequences. Consensus sequences may be identified by sequence comparisons between individual
aptamer species, which comparisons may be aided by computer programs and other tools for modeling secondary and tertiary structure from sequence information. Generally, the consensus sequence will contain at least 5 about 3 to 20 nucleotides, more commonly from 6 to 10 nucleotides.
.As used herein "consensus sequence" means that certain positions, not necessarily contiguous, of an oligonucleotide are specified. By specified is meant 0 that the composition of the position is other than completely random. Not all oligonucleotides in a mixture may have the same nucleotide at such position; for example, the consensus sequence may contain a known ratio of particular nucleotides. For example, a consensus 5 sequence might consist of a series of four positions wherein the first position in all members of the mixture is A, the second position is 25% A, 35% T and 40% C, the third position is T in all oligonucleotides, and the fourth position is G in 50% of the oligonucleotides and C 0 in 50% of the oligonucleotides.
When a consensus sequence is identified, oligonucleotides that contain that sequence may be made by conventional synthetic or recombinant means. These aptamers, termed "secondary aptamers," may also function 5 as thrombin-specific aptamers of this invention. A secondary aptamer may conserve the entire nucleotide sequence of an isolated aptamer, or may contain one or more additions, deletions or substitutions in the nucleotide sequence, as long as a consensus sequence is C conserved. A mixture of secondary aptamers may also function as thrombin-specific aptamers, wherein the mixture is a set of aptamers with a portion or portions of their nucleotide sequence being random or varying, and a conserved region which contains the consensus sequence. Additionally, secondary aptamers may be synthesized using
one or more of the modified bases, sugars and linkages described herein using conventional techniques and those described herein.
Utility of the Aptamers
The aptamers of the invention are useful in diagnostic, research and therapeutic contexts. For therapeutic applications, the thrombin aptamers have in vivo and ____: vivo clinical utilities, as indicated above. By way of example, the aptamers may be used in the treatment or prevention of (i) restenosis or myointimal thickening associated with angioplasty, (ii) accelerated atherosclerosis after heart transplant operations, (iii) vascular graft reocclusion associated with vascular shunt implants, (iv) clotting or thrombus formation at the site of indwelling arterial or venous access lines, (v) thrombus formation associated with cardiopulmonary bypass surgery, (vi) thrombus formation associated with extracorporeal circuits that are used during various ex vivo procedures such as blood dialysis or apheresis, (vii) sepsis-related disseminated intravascular coagulation and (viii) coagulation in patients with known heparin allergy or heparin-indu- 'd thrombocytopenia.
For diagnostic applications, these aptamers are well suited for binding to biomolecules that are identical or similar between different species, where standard antibodies may be difficult to obtain. They are also useful in inhibition assays when the aptamers are chosen to inhibit the biological activity of thrombin. Antibodies are generally used to bind analytes that are detected or quantitated in various diagnostic assays. Aptamers represent a class of molecules that may be used in place of antibodies for in vitro or .in vivo diagnostic and purification purposes.
Aptamers that bind to thrombin may be used as in vivo imaging or diagnostic reagents when suitably radiolabeled. Isotopes such as 131I, 99mTc, 90Y, lι:LIn and J""ι have been used to label various proteins or antibodies as is described in the literature (Cohn, K.H., et al, .Arch. Surg. (1987) 122.:1245-1429; Baidoo, K.E., et al, Cancer Res. (Suppl.) (1990) 5_0_:799s-803s; Beatty, J.D., et al, Cancer Res. (Suppl.) (1990) 50:840s-845: Sharkey, R.M., et al Cancer Res. (1988) 18:32270-3275). A preferred isotope is 99mTc which is utilized as described in the literature. Chemical modifications of oligonucleotides that are compatible with labeling protocols are also known in the art and have been extensively described (Uhlmann, E., et al, Chemical Rev. (1990) 9_0:543-584; international publication Nos. WO 91/14696 and WO 91/13080) .
The thrombin aptamers may also be labelled by linking a moiety that chelates an imaging agent such as 99πTc. In this embodiment, thrombin aptamer would be administered to a patient followed by administration of the imaging agent. In vivo chelation of the imaging agent would occur, allowing subsequent imaging by conventional means.
Thrombin aptamers may also be labeled with contrast agents such as lanthanide or transition metal complexes or nuclei such as 19F, 15N or 32P to facilitate in vivo imaging of clots and similar formations. Imaging would be performed using magnetic resonance imaging techniques known in the art. One consideration in generating radiolabeled antibodies is that the labeling procedure must not destroy its antigen-binding properties. This usually requires an optimized protocol to be generated for each isotope and antibody. Because the aptamers of the
invention are tolerant of harsh chemical conditions, including conditions under which they are synthesized, facile radiolabeling of thrombin aptamers can be conducted without regard to loss of aptamer structure. Only the chemical integrity of the aptamer molecule must be preserved. The aptamers of the invention can be denatured without loss of their capacity to bind thrombin once placed under physiological conditions. Antibodies cannot be reversibly denatured in this manner. Another consideration relevant to the use of monoclonal antibodies (MAbs) for in vivo imaging is their antigenicity. MAbs are usually derived from mouse hybridomas and as such are foreign proteins. When used in humans they elicit immune responses that limits their use in individual patients to one or two exposures. Once immunized, anti-MAb antibodies generated by an immunized individual leads to rapid clearance of the MAb. This consideration is also relevant to "humanized" MAbs that contain both mouse and human protein sequences. In addition to chemical stability, the aptamers described herein have a short half-life, a property that can permit rapid in vivo imaging after administration of labeled compound. The thrombin aptamers can also be advantageously used to avoid anaphylactic reactions such as those associated with imaging procedures that use conventional ionic or nonionic contrast agents. The aptamers also have a low molecular weight compared to .Abs, which can facilitate their penetration of a target structure, such as a clot, for imaging purposes. Radiolabeled thrombin aptamers can be used to image arteries or veins according to various clinical indications. For example, such aptamers can be used after angioplasty to image clots, including deep vein clots, CNS thromboses, pulmonary emboli, brain thromboses and the like.
The aptamers of the invention are therefore particularly useful as diagnostic reagents to detect the presence or absence of thrombin. In vitro diagnostic tests are conducted by contacting a sample with the specifically binding oligonucleotide to obtain a complex which is then detected by conventional means. For example, the aptamers may be labeled using radioactive, fluorescent, or chromogenic labels and the presence of label bound to solid support to which the thrombin has been bound through a specific or nonspecific binding means detected. Alternatively, the specifically binding oligonucleotides may be used to effect initial complexation to the support. Means for conducting assays using such oligomers as specific binding partners will track those for standard specific binding partner based assays.
It may be commented that the mechanism by which the specifically binding oligomers of the invention interfere with or inhibit the activity of thrombin is not always established, and is not a part of the invention.
The oligomers of the invention are characterized by their ability to bind thrombin regardless of the mechanisms of binding or the mechanism of the effect thereof.
For use in research or manufacturing, the specifically binding oligonucleotides of the invention are especially helpful in effecting the isolation and purification of substances to which they bind. For this application, typically, the aptamer containing the specific binding sequences is conjugated to a solid support and used as an affinity ligand in chromatographic separation of thrombin.
In therapeutic applications, the aptamers of the invention can be formulated for a variety of modes of administration, including systemic and topical or localized administration. Techniques and formulations
generally may be found in Remington's Pharmaceutical Sciences. Mack Publishing Co., Easton, PA, latest edition. In general, the dosage required for therapeutic efficacy will range from about 0.1 μg to 20 mg aptamer/kg body weight. Alternatively, dosages within these ranges can be administered by constant infusion over an extended period of time, usually exceeding 24 hours, until the desired therapeutic benefits have been obtained.
For systemic administration, injection is preferred, including intramuscular, intravenous, intraperitoneal, and subcutaneous. For injection, the aptamers of the invention are formulated in liquid solu¬ tions, preferably in physiologically compatible buffers such as Hank's solution or Ringer's solution. In addi- tion, the aptamers may be formulated in solid form and redissolved or suspended immediately prior to use. Lyophilized forms are also included.
Systemic administration can also be by transmucosal or transdermal means, or the oligomers can be administered orally. Additional formulations which are suitable for other modes of administration include suppositories, intranasal and other aerosols. For transmucosal or transdermal administration, penetrants appropriate to the barrier to be permeated are used in the formulation. Such penetrants are generally known in the art, and include, for example, for transmucosal administration bile salts and fusidic acid derivatives. In addition, detergents may be used to facilitate permeation. Transmucosal administration may be through nasal sprays, for example, or using suppositories. For oral administration, the oligomers are formulated into conventional oral administration forms such as capsules, tablets, and tonics.
For topical administration, the oligomers of the invention are formulated into ointments, salves, gels, or creams, as is generally known in the art.
The aptamers may also be employed in expression systems, which are administered according to techniques applicable, for instance, in applying gene therapy.
The following examples are meant to illustrate, but not to limit the invention.
Selection of Aptamers that Bind to Thrombin
A. Synthesis of Oligonucleotide Pool
DNA oligonucleotides containing a randomized sequence region were synthesized using standard solid phase techniques and phosphoramidite chemistry (Oligonucleotide Synthesis. Gait, M.J., ed. (IRL Press), 1984; Cocuzza, A., Tetrahedron Letters. (1989) 30.:6287- 6291) . A 1 μM small-scale synthesis yielded 60 nmole of HPLC-purified single-stranded randomized DNA. Each strand consisted of specific 18-mer sequences at both the 5' and 3' ends of the strand and a random 60-mer sequence in the center of the oligomer to generate a pool of 96-mers with the following sequence (N = G, A, T or C) :
5' HO-CGTACGGTCGACGCTAGCNgQCACGTGGAGCTCGGATCC-OH 3
DNA 18-mers with the following sequences were used as primers for PCR amplification of oligonucleotide sequences recovered from selection columns. The 5' primer sequence was 5' HO-CGTACGGTCGACGCTAGC-OH 3' and the 3' primer sequence was 5' biotin-O-
GGATCCGAGCTCCACGTG-OH 3'. The biotin residue was linked to the 5' end of the 3' primer using commercially available biotin phosphoramidite (New England Nuclear,
Cat. No. NEF-707) . The biotin phosphoramidite is incorporated into the strand during solid phase DNA synthesis using standard synthesis conditions.
In another, similar experiment, a pool of 98-mers with the following sequence was synthesized:
5' H0-AGAATACTC^AGCTTGCCG-N60-ACCTGAATTCGCCCTATAG-OH 3'.
DNA 19-mers with the following sequences can also be used as primers for PCR amplification of oligonucleotides recovered from selection columns. The 3' primer sequence is
5' biotin-0-CTATAGGGCGAATTCAGGT-OH 3'
and the 5' primer sequence is
5' HO-AGAATACTCAAGCTTGCCG-OH 3'.
It will be noted that in all cases, the duplex form of the primer binding sites contain restriction enzyme sites.
B. Isolation of Thrombin Aptamers Using Thrombin Immobilized on a Lectin Column
A pool of aptamer DNA 96 bases in length was synthesized as described in Example 1-A, and then PCR- amplified to construct the initial pool. A small amount of the enzymatically-synthesized DNA was further amplified in the presence of α- 32P-dNTPs to generate labeled aptamer to permit quantitation from column fractions.
A thrombin column was prepared by washing 1 mf
(58 nmole) agarose-bound concanavalin A ("Con-A") (Vector Laboratories, cat. no. AL-1003) with 20 mM Tris-acetate
buffer (pH 7.4) containing 1 mM MgCl2, 1 mM CaCl2, 5 mM KC1 and 140 mM NaCl (the "selection buffer") (4 x 10 mf) . 1 mf of settled support was then incubated overnight at 4°C in 10 mf selection buffer containing 225 μg (6.25 nmole) thrombin (Sigma, Cat. no. T-6759) . After shaking overnight to permit thrombin binding to the Con-A beads, the mixture was briefly centrifuged and the supernatant removed. The beads were resuspended in fresh selection buffer and transferred to a column which was then washed with selection buffer (5 x 1 mf ) . A column containing 1 mf of settled beads had a void volume of approximately 300 μL. A control Con-A column was prepared by adding 1 mf of settled support to a column followed by 5 washes of 1 mf of selection buffer. Prior to application of the aptamer DNA pool to
Con-A columns, the DNA was heated in selection buffer at 95°C for 3 minutes and then cooled on ice for 10 minutes. The pool, consisting of 100 pmole DNA in 0.5 mf selection buffer, was then pre-run on the control Con-A column at room temperature to remove species that bound to the control support. Three additional 0.5 mf aliquots of selection buffer were added and column fractions 2, 3 and 4 (0.5 mf each) were pooled and then reapplied to the column twice. The DNA in 1.5 mf selection buffer was then recovered. Approximately 1% of total input cpm were retained on the column.
The recovered DNA was then applied to a Con-A- thro bin column as a 0.5 mf aliquot followed by a 1.0 mf aliquot. Flow-through was retained and reapplied to the column twice. DNA added to the column on the final application was left on the column for 1 hour at room temperature. The column was then eluted with 0.5 mf aliquots of selection buffer. 0.5 mf fractions were collected and radioactivity was determined in each fraction. Radioactivity in eluted fractions 7 through 12
were low and relatively constant. After recovery of fraction 12, the column was washed with 0.5 mf aliquots of 0.1 M α-methyl-mannoside (Sigma Cat. no. M-6882) in selection buffer to elute the bound thrombin along with thrombin-bound aptamers. Fractions 14 and 15 showed a significant peak of thrombin enzyme activity, as determined spectrophotometrically by conversion of a chromogenic substrate (Kabi Diagnostica, Cat. no. S-2238) . 0.01% of the input DNA eluted in these two fractions.
Aptamer DNA (Round 1 DNA) was recovered from the thrombin by phenol extraction (2 x 0.5 m ) . The aqueous phase volume was reduced to about 250 μl by n- butanol extraction. Aptamer DNA was precipitated on dry ice using 3 volumes of ethanol and 20 μg of glycogen as a carrier. The DNA was pelleted, washed once in 70% ethanol and then dried.
C. Amplification of Selected Thrombin Aptamers Round 1 DNA from Example 1-B was resuspended in
100 μl of H20 and amplified by PCR. A 200 μl PCR reaction consisted of the following: 100 μl template 96- mer DNA (approximately 0.01 pmoles) ; 20 μl 10X buffer (100 mM Tris-Cl (pH 8.3), 500 mM KCl, 20 mM MgCl2) ; 32 μl dNTP's (5 mM cone total, 1.25 mM each dATP, dCTP, dGTP, and dTTP) ; 20 μl primer 1 (biotinylated 18-mer, 50 μM) ; 20 μl primer 2 (18-mer, 50 μM) ; 6μl α-32P-dNTP's (approximately 60 μCi) ; and 2 μl Taq I Polymerase (10 units) . The reaction was covered with 2 drops NUJOL mineral oil. A control reaction was also performed without template aptamer.
Initial denaturation was at 94°C for 3 minutes, but subsequent denaturation after each elongation reaction lasted 1 minute. Primer annealing occurred at 60°C for 1 minute, and elongation of primed DNA strands
using the Taq polymerase ran at 72°C for 2 minutes, with 5-second extensions added at each additional cycle. The final elongation reaction to completely fill in all strands ran for 10 minutes at 72°C, and the reaction was then held at 4°C.
18 rounds of Taq polymerase elongation were carried out in order to amplify the selected aptamer DNA. After the reactions were completed, the aqueous layer was retrieved and any residual NUJOL oil was removed by n- butanol extraction, reducing the volume to 100 μL. A sample may be removed from each of the aptamer and control reaction for quantitation and analytical PAGE. The amplified aptamer pool (100 μL) was run over a Nick column (G-50 Sephadex, washed with 3 mL TE buffer (10 mM Tris-HCl (pH 7.6), 0.1 mM EDTA)) to remove unincorporated NTP's, primers, and salt. 400 μL of TE buffer was then added to the column and the DNA pool was eluted from the column with an additional 400 μL using TE buffer. (A sample may be removed from the eluent for quantitation and analytical PAGE.) The eluent (400 μL) was loaded on an avidin agarose column (Vector Laboratories, Cat. No. A-2010) (500 μL settled support, washed with 3 x 1 mL Tris/NaCl buffer (0.1 M Tris, 0.1 M NaCl, pH 7.5)) . Approximately 90% of the loaded radioactivity remained on the column. The column was washed with Tris/NaCl buffer
(4 x 400 μl) and then the nonbiotinylated strand was eluted with 0.15 N NaOH (3 x 300 μL fractions) . More than 45% of the radioactivity on the column eluted in these three fractions. These fractions (900 μl) were combined and neutralized with approximately 3.5 μl of glacial acetic acid. The neutralized fractions were reduced to 250 μl by speed vacuum or butanol extraction and the nucleic acids were precipitated with EtOH. The resultant pellet was dissolved in 102 μl selection buffer. A 2 μl sample was removed for quantitation and
analytical PAGE. The resulting amplified Round 1 Pool was applied to a new Con-A-thrombin column as in Example l-B to obtain Round 2 aptamers.
D. Characterization of Round 1 Through Round 5 Thrombin Aptamers Obtained from Selection on Lectin Columns Five rounds of thrombin aptamer selection and amplification were carried out using Con-A-thrombin columns as in Examples l-B and 1-C. As shown in Table l, the combined fractions 14 and 15 contained a maximum of about 10% of input DNA using the described conditions.
Table 1
% DNA eluted by # % DNA bound to
Round α-methyl-mannoside control support
1 0.01 0.7
2 0.055 1.9
3 5.80 2.3 4 10.25 1.1
5 9.70 1.0
0.1 M α-methyl-mannoside in selection buffer was added as fraction 13 in each elution, and fractions 14 and 15 were retained and the DNA amplified. Due to slow leeching of thrombin from the column, DNA bound to thrombin could also be seen in earlier fractions in rounds 3-5.
After amplification, round 5 aptamer DNA was analyzed for specificity in a filter binding assay. In this assay, nitrocellulose filters (1 cm diameter) prebound with salmon sperm DNA were used to bind either: (l) An unselected 96-mer oligonucleotide DNA pool, (2)
unselected DNA with thrombin (60 pmole) , (3) Round 5 aptamer DNA and thrombin (60 pmole) , (4) Round 5 aptamer DNA alone, or (5) Round 5 aptamer DNA and ovalbumin (60 pmole) . In each case 3.5 pmole of DNA was used and the 5 incubation was in 200 μL selection buffer at room temperature for 1 hour. The filters were then washed 3 times with 3.0 mf of selection buffer and radioactivity was counted to determine the amount of DNA that was retained as a thrombin complex. The results are shown in 0 Table 2.
Table 2
DNA % DNA Bound to Filter 5
Unselected 96-mer 0.08
Unselected 96-mer + thrombin 0.06
Round 5 aptamer + thrombin 20.42
Round 5 aptamer 0.07 0 Round 5 aptamer + ovalbumin 0.05
Unselected DNA did not show significant binding to the thrombin while selected aptamer DNA bound to ς thrombin. Binding results show specific thrombin binding with no detectable ovalbumin binding.
Round 5 aptamer DNA was then amplified using the following 3' primer sequence:
0 5' HO-TAATACGACTCACTATAGGGATCCGAGCTCCACGTG-OH 3
and the 5' 18-mer primer sequence shown in Example l-A. The 36-mer primer was used to generate internal BamHl restriction sites to aid in cloning. The amplified Round 5 aptamer DNA was then cloned into pGEM 3Z (Promega) . 32
of the resulting clones were then amplified directly using the following 5' primer sequence:
5' HO-CTGCAGGTCGACGCTAGC-OH 3'
and the 3' biotinylated 18-mer primer sequence shown in Example 1-A, and then sequenced.
Filter binding assays using aptamer DNA from 14 of the clones were used to determine the dissociation constants (Kd) for thrombin as follows: Thrombin concentrations between 10 μM and 1 nM were incubated at room temperature in selection buffer for 5 minutes in the presence of 0.08 pmole of radiolabeled 96-mer derived from cloned Round 5 aptamer DNA. After incubation, the thrombin and aptamer mixture was applied to nitrocellulose filters (0.2 micron, 2.4 cm diameter) that were pretreated with salmon sperm DNA (1 mg/mf DNA in selection buffer) and washed twice with 1 mf selection buffer. After application of thrombin mixture, the filters were washed three times with 1 mf selection buffer The radioactivity retained on the filters was then determined. Kd values for the individual clones ranged from 50 to >2000 nM.
The DNA sequence of the 60-nucleotide randomly- generated region from 32 clones was determined in order to examine both the heterogeneity of the selected population and to identify homologous sequences. Sequence analysis showed each of the 32 clones to be distinct. However, striking sequence conservation was found. The hexamer 5' GGTTGG 3' was found at a variable location within the random sequence in 31 of 32 clones, and five of the six nucleotides are strictly conserved in all 32. Additionally, in 28 of the 32 clones a second hexamer 5' GGNTGG 3', where N is usually T and never C,
is observed within 2-5 nucleotides from the first hexamer. Thus, 28 clones contain the consensus sequence 5' GGNTGG(N)ZGGNTGG 3' where z is an integer from 2 to 5. The remaining 4 clones contain a "close variant sequence" (a sequence differing by only a single base) . A compilation of the homologous sequences are shown in Figure l. It should be noted that DNA sequencing of several clones from the unselected DNA population or from a population of aptamers selected for binding to a different target revealed no homology to the thrombin- selected aptamers. From these data we conclude that this consensus sequence contains a sequence which is responsible either wholly or in part, for conferring thrombin affinity to the aptamers. Clotting time for the thrombin-catalyzed conversion of fibrinogen (2.0 mg/mL in selection buffer) to fibrin at 37°C was measured using a precision coagulation timer apparatus (Becton-Dickinson, Cat. nos. 64015, 64019, 64020) . Thrombin (10 nM) incubated with fibrinogen alone clotted in 40 sec, thrombin incubated with fibrinogen and PI nuclease (Boehringer-Mannheim, Indianapolis, IN) clotted in 39 sec, thrombin incubated with fibrinogen and aptamer clone #5 (200 nM) clotted in 115 sec, and thrombin incubated with fibrinogen, clone #5 (200 nM) and PI nuclease clotted in 40 sec. .All incubations were carried out at 37°C using reagents prewarmed to 37°C. Aptamer DNA or, when present, PI nuclease, was added to the fibrinogen solution prior to addition of thrombin. These results demonstrated that ;i) thrombin activity was inhibited specifically by intact aptamer DNA and (ii) that inhibitory activity by aptamer did not require a period of prebinding with thrombin prior to mixing with the fibrinogen substrate.
Inhibition of thrombin activity was studied using a consensus-related sequence 7-mer, 5' GGTTGGG 3',
or a control 7-mer with the same base composition but different sequence (5' GGGGGTT 3'). Clotting times were measured using the timer apparatus as above. The thrombin clotting time in this experiment was 24 sec using thrombin alone (10 nM) , 26 sec with thrombin and the control sequence at 20 μM and 38 sec with thrombin plus the consensus sequence at 20 μM, indicating specificity for thrombin inhibition at the level of the 7-mer. The inhibitory aptamers were active at physiological temperature under physiologic ion conditions and were able to bind to thrombin in the presence of the fibrinogen substrate, a key requirement for therapeutic efficacy.
Example 2 Modified Thrombin Aptamers Modified forms of the single-stranded, thrombin consensus sequence-containing deoxynucleotide 15-mer described in Example 2, 5' GGTTGGTGTGGTTGG 3', and a closely related 17-mer, were synthesized using conventional techniques. These aptamers for the most part contain the identical nucleotide sequences, bases, sugars and phosphodiester linkages as conventional nucleic acids, but substitute one or more modified linking groups (thioate or MEA) , or modified bases (uracil or 5- (i-pentynyl-2' -deoxy)uracil) . The aptamers containing 5- (1-pentynyl) -2' -deoxyuridine were generated by replacing thymidine in the parent aptamers. Thrombin aptamers containing 5- (1-pentynyl) -2' -deoxyuridine were also obtained by selection as described in Examples 8 and 9 below.
Independent verification of the Ki for the nonmodified 15-mer was made by determining the extent of thrombin inhibition with varying DNA concentration. The
data revealed 50% inhibition of thrombin activity at approximately the same concentration as the derived Ki, strongly suggesting that each bound thrombin was largely, if not completely, inhibited, and that binding occurred with a 1:1 stoichiometry.
Table 3 Compound Ki (nM)
GGTTGGTGTGGTTGG 20
GGTTGGTGTGGTTGG#G#T 35
GGTTGGTGTGGTT*G*G 40
G*G*T*T*G*G*T*G*T*G*G*T*T*G*G 280
GGTTGG(dU)G(dU)GGTTGG 15 GG(dU)TGGTGTGG(dU)TGG 80
GGTTGGTGTGGTU'GG 20
indicates a thioate (i.e., P(O)S) linkage * indicates a MEA linkage U' indicates 5- (l-pentynyl)uracil
Example 3 Incorporation of 5- (l-pentynyl) -2' -deoxyuridine Into Aptamer Candidate DNA 5- (l-pentynyl) -2' -deoxyuridine was synthesized and converted to the triphosphate as described in Otvos, L., et al., Nucleic Acids Res (1987) 1763-1777. The pentynyl compound was obtained by reacting 5-iodo-2'- deoxyuridine with 1-pentyne in the presence of palladium catalyst. 5- (l-pentynyl) -2' -deoxyuridine triphosphate was then used as a replacement for thymidine triphosphate in the standard PCR reaction.
A pool of 96-mer single-stranded DNA was synthesized, each strand consisting of specific 18-mer
PCR primer sequences at both the 5' and 3' ends and a random 60-mer sequence in the center of the oligomer. Details of synthesis of the pool of single-stranded DNA is disclosed in Example 1 above. PCR conditions were the same as those described above, with the following changes. dATP, dGTP and dCTP were all used at a concentration of 200 μM. The optimal concentration for synthesis of full-length 96-mer DNA via PCR using 5- (1- pentynyl) -2' -deoxyuridine was 800 μM. Generation of PCR- amplified fragments demonstrated that the Taq polymerase both read and incorporated the base as a thymidine analog. Thus, the analog acted as both substrate and template for the polymerase. Amplification was detected by the presence of a 96-mer band on an EtBr-stained polyacrylamide gel.
Example 4 Incorporation of Other Base .Analogs Into Candidate Aptamer DNA Ethyl, propyl and butyl derivatives at the
5-position of uridine, deoxyuridine, and at the N -position of cytidine and deoxycytidine are synthesized using methods described above. Each compound is converted to the triphosphate form and tested in the PCR assay described in Example 1 using an appropriate mixture of three normal deoxytriphosphates or ribotriphosphates along with a single modified base analog.
This procedure may also be performed with N -position alkylated analogs of adenine and deoxyadenine, and the 7-position alkylated analogs of deazaguanine, deazadeoxyguanine, deazaadenine and deazadeoxyadenine, synthesized using methods described in the specification.
Example 5 Thrombin Aptamer Containing Substitute
Internucleotide Linkages Modified forms of the 15-mer thrombin aptamer, 5' GGTTGGTGTGGTTGG 3' containing one or two formacetal internucleotide linkages (0-CH2-0) in place of the phosphodiester linkage (O-PO(O')-O) were synthesized and assayed for thrombin inhibition as described above. The H-phosphonate dimer synthon was synthesized as described in Matteucci, M.D., Tet. Lett. (1990) 31:2385-2387. The formacetal dimer, 5' T-0-CH2-0-T 3', was then used in solid phase synthesis of aptamer DNA. Control unmodified aptamer DNA was used as a positive control. The results that were obtained are shown in Table 4.
Table 4 Compound clot time (sec)
100 nM 20 nM 0 nM
GGT TGGTGTGGTTGG 105 51
GGTTGGTGTGGT TGG 117 48
GGT TGGTGTGGT TGG 84 60
GGTTGGTGTGGTTGG 125 49
NO DNA CONTROL -- -- 25
indicates a formacetal linkage
Example 6 Thrombin Aptamer Containing Abasic Nucleotide Residues
Modified forms of the 15-mer thrombin aptamer, 5' GGTTGGTGTGGTTGG 3' containing one abasic residue at each position in the aptamer were synthesized and assayed for thrombin inhibition as described above. The abasic residue, l,4-anhydro-2-deoxy-D-ribitol was prepared as
described in Eritja, R. , et al, Nucleosides and Nucleotides (1987) 6 :803-814. The N,N-diisopropylamino cyanoethylphosphoramidite synthon was prepared by standard methods as described in Caruthers, M.H. Accounts 5 Chem. Res. (1991) 24:278-284, and the derivatized CGP support was prepared by the procedures described in Dahma, M.J., et al, Nucleic Acids Res. (1990) 18:3813. The abasic residue was singly substituted into each of the 15 positions of the 15-mer. Control unmodified 10 aptamer DNA was used as a positive control. The results that were obtained are shown in Table 5.
15
20
25
30
X - indicates an abasic residue
Example 7
Thrombin Aptamers Containing 5- (1-Propyny1) -2'-deoxyuridine Nucleotide Residues
Modification of the 15-mer thrombin aptamer, 5' GGTTGGTGTGGTTGG 3' to contain 5- (1-propynyl) -2'- deoxyuridine nucleotide analogs at the indicated positions in the aptamer was effected by the synthesis of these aptamers. They were assayed for thrombin inhibition as described above. The aptamer and the H-phosphonate were prepared as described in DeClercq, E., et al, J. Med.Chem. (1983) 26_:661-666; Froehler, B.C., et
al, Nucleosides and Nucleotides (1987) 6.:287-291; and Froehler, B.C., et al, Tet. Lett. (1986) 2.7:469. This analog residue was substituted at the indicated portions and the aptamer assayed for inhibition of thrombin. The results that were obtained are shown in Table 6.
Z - indicates a 5-propynyl-2' -deoxyuridine residue
Example 8 Incorporation of 5- (1-pentvnyl) -2' -deoxyuridine Into Aptamer Candidate DNA 5- (l-pentynyl) -2' -deoxyuridine was synthesized and converted to the triphosphate as described in Otvos, L., et al., Nucleic Acids Res (1987) 1763-1777. The pentynyl compound was obtained by reacting 5-iodo-2'- deoxyuridine with 1-pentyne in the presence of a palladium catalyst. 5- (l-pentynyl) -2' -deoxyuridine triphosphate was then used as a replacement for thymidine triphosphate in the standard PCR reaction.
A pool of 60-mer single-stranded DNA was synthesized, each strand consisting of specific 18-mer PCR primer sequences at both the 5' and 3' ends and a
rando 20-mer sequence in the center of the oligomer. Details of synthesis of the pool of single-stranded DNA is disclosed in Example 1.
Because of the poor substrate activity of pentynyl dUTP when used with TAQ polymerase, VENT™ thermostable polymerase, (New England Biolabs, Cat. No. 254) was employed. Amplification was performed as per the manufacturers instructions. Pentynyl dUTP was included in the reaction as a substitute for dTTP. The single-stranded 60-mer was isolated by a modification of standard procedures. The 200 μL PCR amplification reaction was divided into two samples which were applied to two NICK™ columns equilibrated (5 mL) as described. The eluent was collected, pooled and applied to avidin- agarose as described. This column was washed with buffer followed by elution of single-stranded 60-mer DNA with 0.15 N NaOH, pooled and neutralized with glacial acetic acid. Single-stranded 60-mer DNA was desalted on a NAP5 column equilibrated in 20 mM Tris OAc (pH 7.4) . 10X selection buffer salts were added to the sample, heated to 95°C for 3 minutes, and transferred to wet ice for 10 minutes.
Example 9 Isolation of Thrombin Aptamers Using
DNA Containing 5- (1-Pentynyl) -2'-deoxyuridine
The pool of aptamer DNA 60 bases in length was used essentially as described in Example 8. The aptamer pool sequence was 5' TAGAATACTCAAGCTTCGACG-N20-AGTTTGGATCCCCGGGTAC 3', while the 5' primer sequence was 5'TAGAATACTCAAGCTTCGACG 3' and the 3' biotin-linked primer was 5' GTACCCGGGGATCCAAACT 3' .
Thrombin immobilized on a Con-A lectin column served as the target as described.
After five rounds of selection, aptamer DNA was recovered and amplified using thymidine triphosphate (dTTP) in place of 5- (l-pentynyl) -2' -deoxyuridine in order to facilitate subsequent cloning and replication of aptamer DNA in E__ coli. At this stage, the presence of a thymidine nucleotide at a given location in an aptamer corresponded to the location of a 5- (l-pentynyl) -2' - deoxyuridine nucleotide in each original round five aptamer. Thus, dTTP served to mark the location of 5- (l-pentynyl) -2' -deoxyuridine residues in the original selected DNA pools.
The round five amplified DNA containing dTTP was digested with BamHI and Hindlll and cloned into the corresponding sites of pGEM 3Z (Promega Biotech) and transformed into E. coli. DNA from 21 clones was analyzed by dideoxy sequencing. Three of the clones contained aptamer sequences that were identical. Only one of the 21 clones contained a sequence that closely resembled the original 5' GGTTGG 3' binding motif obtained using thymine in the selection protocol.
One of these two clones (#17) and the original unselected pool was analyzed for thrombin binding by nitrocellulose filter assay described above using DNA labeled with 32P to permi.t analysis of thrombi.n binding characteristics. The labeled DNA was synthesized by PCR and contained 5- (l-pentynyl) -2' -deoxyuridine in order to retain the original selected DNA structures. The DNA was incubated with thrombin at various concentrations between 10 nM and 10 μM to obtain the Kd values for thrombin binding. The Kd of the unselected pool was >10 μM while the Kd of clone 17 was 300 nM.
Radiolabeled clone 17 DNA was synthesized using thymidine in place of 5- (l-pentynyl) -2' -deoxyuridine and
the resulting DNA had a Kd of >10 μM, demonstrating that the 5- (l-pentynyl) -2'-deoxyuracil heterocycle could not be replaced by thymine in the selected aptamer without loss of binding affinity. 5 Representative sequences that were obtained are as follows.
5' TAGTATGTATTATGTGTAG 3'
5' ATAGAGTATATATGCTGTCT 3'
5' GTATATAGTATAGTATTGGC 3' 10 5' AGGATATATGATATGATTCGG 3'
5' TACTATCATGTATATTACCC 3'
5' CATTAAACGCGAGCTTTTTG 3'
5' CTCCCAT.AATGCCCTAGCCG 3'
5' GACGCACCGTACCCCGT 3' 15 5' CACCAAACGCATTGCATTCC 3'
5' GTACATTCAGGCTGCCTGCC 3'
5' TACCATCCCGTGGACGTAAC 3'
5' GACT.AAACGCATTGTGCCCC 3'
5' AACGAAGGGCACGCCGGCTG 3' 20 5' ACGGATGGTCTGGCTGGACA 3'
Example 10 Isolation of Thrombin Aptamers Using 25 DNA Containing 5-Methyl -2 '-deoxycytidine
5-methyl-2'-deoxycytidine triphosphate was obtained commercially (Pharmacia, Cat. No. 27-4225-01) and used to synthesize DNA containing random sequences 60 bases in length flanked by primers 19 bases in length. 3C The pool of aptamer DNA 98 bases in length was used essentially as described in Example 1. Thrombin immobilized on a Con-A lectin column ser-ved as the target as described.
Briefly, a 200 μL PCR reaction was set up
■3C using: 10 mM Tris-HCl, pH 8.3 at 25° C, 1.5 mM MgCl , 5C
mM NaCl and 200 μM of each of dATP, dGTP, dTTP and 5- methyl-2' -deoxycytidine triphosphate. 20 μCi each of α- P-dATP and dGTP were added to label the DNA. 1 nmole of 5' and 3' primer were added followed by addition of 0.2 pmole of 98-mer template pool DNA. Amplification was initiated by addition of 2 μL (10 U) of Taq polymerase followed by sealing of the reaction with a mineral oil overlay. About 16 cycles of amplification were performed followed by a 10 minute final extension to complete all duplex synthesis.
Amplified DNA was recovered (100 μL aqueous phase) , n-butanol extracted (650 μL) and applied to a Nick column prewashed with 5 mL of buffer containing 100 mM Tris-HCl pH 7.5 and 100 mM NaCl. Eluted DNA was applied to a 0.5 mL avidin-agarose column prewashed in the same buffer and washed until DNA loss from the column was < 1000 cpm. Single stranded DNA was eluted from the avidin column by washing with 0.15 N NaCl and the eluate was neutralized to pH 7.0 using glacial acetic acid. The 98-mer DNA was exchanged into selection buffer on a second Nick column and, after heat denaturation for 3 min at 95° C followed by cooling on ice for 10 min, used in aptamer selection on thrombin lectin columns. 1 mL thrombin columns were equilibrated in selection buffer prior to addition of single-stranded DNA. The single- stranded DNA was recirculated for three complete passes. Upon completion of the third pass the peak radioactive element was then applied to a 1 mL ConA/thrombin column (charged with 3 nmoles of thrombin) . Radioactive single- stranded 98-mer was applied three times to this matrix. At the third application, the column was stoppered and allowed to stand for 1 hr. The column was then washed with selection buffer and 0.5 mL aliquot fractions collected. A total wash volume of 6 mL was employed. At this time, 0.1 M oc-methyl-mannoside in selection buffer
was then added, followed by a 4 mL total volume wash. Thrombin enzymatic activity was detected via chromogenic substrate monitored by absorbance at 405 nm. Peak thrombin fractions were pooled, extracted with phenol, and the volume reduced by nBuOH extraction. 20 μg glycogen was added, the single-stranded 98-mer precipitated via ethanol addition and pelleted via centrifugation. The pelleted DNA was resuspended in water and used as a template for PCR amplification. This protocol was repeated to obtain a pool of DNA that resulted from 5 rounds of selection on thrombin columns.
Double-stranded DNA was digested with EcoRI and HinDIII and cloned into pGEM3Z. Aptamers were then transformed into E. coli and analyzed by dideoxy sequencing. Round five aptamer pool DNA bound to thrombin with a Kd of approximately 300 nM.
Example 11 Demonstration of Aptamer Specificity for Binding tn and Inhibition of Thrombin
The specificity of aptamer binding was demonstrated using 32P radiolabeled DNA and a series of proteins. To determine the binding specificity of the thrombin aptamer, 96-mer clone #29, having the partial sequence 5'CGGGGAGAGGTTGGTGTGGTTGGC.AATGGCTAGAGTAGTGAC GTTTTCGCGGTGAGGTCC 3' was used. The consensus sequence is shown underlined. In addition, a 21-mer aptamer, 5' GGTTGGGCTCGTTGGGTTGGG 3' was tested for inhibition of another fibrinogen-cleaving enzyme ancrod, which was obtained commercially (Sigma, Cat. No. A-5042) . The
21-mer had a of Ki for thrombin of about 100 nM and its Kd was about 350 nM. Clone #29 had a Kd of about 200 nK for thrombin.
The aptamer was shown to specifically bind to thrombin by a filter binding assay. Briefly,
radiolabeled aptamer DNA at.about a concentration of about 1 nM was incubated with the indicated protein for several minutes at room temperature, followed by filtration of the aptamer-protein mixture through a nitrocellulose filter. The filter was washed with 3 mL of selection buffer and then radioactivity bound to the filters was determined as a % of input radioactivity. Results obtained are shown in Table 7. Binding data is shown for both unselected 96-mer DNA and for two separate experiments with clone #29 96-mer. All proteins were tested at about lμM concentration except human serum albumin which was used at 100 μM. The results that were obtained demonstrated that the 96-mer specifically bound to thrombin and had little affinity for most of the other proteins tested.
Bound
0
5 0 59.0 11.0 2.0 <0.5 0 <0.5 <0.5 15. C <0.5
5
0 59.0 9.5 <0.5 C <0.5 <0.5 <0.5 15. C
<0.5
The thrombin 21-mer ancrod assay was conducted as follows. Ancrod was suspended in sterile water at a concentration of 44 U/mL. 10 μL ancrod solution was added to 95 μL of selection buffer prewarmed to 37°C. 100 μL of this mixture was transferred to the coagulation cup of the fibrometer described above, followed by addition of 200 μL of fibrinogen and 20 μL of 21-mer DNA (both prewarmed to 37°C) . TE buffer pH 7.0 was used as a control lacking DNA. The control clot time was 25 seconds while the clot time in the presence of 500 nM 21- mer was 24 seconds and was 26 seconds in the presence of 33 μM 21-mer. This result demonstrated the specificity on inhibition of fibrinogen cleavage was limited to thrombin; ancrod was not affected.
Example 12 Thrombin Aptamer Pharmacokinetic Studies A 15-mer single-stranded deoxynucleotide, 5' GGTTGGTGTGGTTGG 3', identified as a consensus sequence from 30 thrombin aptamer clones as described in Example 1 above, was used. Young adult rats of mixed gender and strain were used. The animals were anaesthetized and a diester of the 15-mer was injected through a catheter in 200 μl volumes (in 20 mM phosphate buffer, pH 7.4, 0.15 M NaCl) at two concentrations, so that the final concentration of 15-mer in the blood was about 0.5 and 5.0 μM respectively, although the exact concentration depends on the volume of distribution (which is unknown for this oligonucleotide) . These values are 10 to 100 times greater than the human in vitro Kd value. No heparin was used for catheterization.
At 0, 5, 20 and 60 minutes, blood was withdrawn from the animals (approx. 500 μl aliquots) , transferred into tubes containing 0.1 volume citrate buffer, and centrifuged. Rat plasma was removed and tested in a
thrombin clotting-time assay. Six animals were used at each concentration, and three animals were injected with the control carrier solution containing no 15-mer.
A prolonged clotting time was observed at the 5 minute time point at both concentrations, with the most significant prolongation occurring at the higher dose concentration. Little or no activity was observed at 20 minutes. Thus, the 15-mer in blood withdrawn from rats 5 minutes post-injection was able to inhibit exogenously added human thrombin. A separate APTT test at the .5 minute time point showed that the 15-mer also inhibited rat blood coagulation, presumably by inhibiting rat thrombin to a significant degree. The half-life of the 15-mer in rats appears to be about 2 minutes or less.
Example 13 Thrombin Aptamer Primate Studies Two thrombin aptamers were administered to adult male cynomologous monkeys. Unsubstituted 15-mer DNA with the sequence 5' GGTTGGTGTGGTTGG 3' and an analog, 5' GGTTGGTGTGGTT*G*G 3', containing thioate internucleotide linkages at the indicated positions (*) , were used. Aptamer was delivered as an intravenous bolus or infusion and then blood samples were withdrawn at various times after delivery of the bolus or during and after infusion. The catheter was heparinized after the 10 minute timepoint. The animals were not systematically heparinized.
Thrombin inhibition was measured by a prothrombin time test (PT) using a commercially available kit, reagents and protocol (Sigma Diagnostics, St. Louis, catalog Nos. T 0263 and 870-3). Inhibition of thrombin was indicated by an increased clot time compared to the control in the PT test. Clot times were obtained by withdrawing a sample of blood, spinning out red cells and
using the plasma in the PT test. Control thrombin PT clot time values were obtained several minutes prior to administration of aptamer. Briefly, the PT assay was conducted using 0.1 mL of monkey plasma prewarmed to 37° C and 0.2 mL of a 1:1 mixture of thromboplastin (used according to manufacturers instructions) and CaCl2 (25 mM) , also prewarmed to 37°C. Thrombin clot times were measured with a fibrometer as described above.
The animals were at least two years old and varied in weight from 4 to 6 kg. Doses of aptamer were adjusted for body weight. Aptamer DNA was dissolved in sterile 20 mM phosphate buffer (pH 7.4) at a concentration of 31.8 to 33.2 mg/mL and diluted in sterile physiological saline prior to delivery. Bolus injections were administered to give a final concentration of 22.5 mg/Kg (1 animal) of the diester aptamer or 11.25 mg/Kg (1 animal) of the diester aptamer. Infusions were administered over a 1 hour period to three groups of animals: (i) 0.5 mg/kg/min of diester 15-mer (4 animals), (ii) 0.1 mg/kg/min of diester 15-mer (2 animals) and (iii) 0.5 mg/kg/min of thioate analog 15- mer (2 animals) .
PT assay results from the bolus injections showed thrombin inhibition times of 7.8, 3.3 and 1.35 times control at 2.5, 5.0 and 10.0 min respectively after delivery of the aptamer for the high dose animal. Inhibition times of 5.6, 2.2 and 1.2 times control were obtained from the low dose animal at the same time points. Figure 2 shows a plot of the PT times from the
4 animals that received the high dose diester infusion compared to pretreatment control values. The data points show the PT clot time as an average value obtained from the 4 animals in the group. The arrows indicate time points at the beginning and end of the infusion period.
Thrombin inhibition peaked at about 10 to 20 min after the infusion was initiated and remained level until the infusion period was terminated. Inhibitory activity decreased rapidly after the infusion of aptamer terminated.
High dose diester and high dose thioate animals showed comparable inhibition of thrombin-mediated clotting, with the high dose thioate giving a sustained clot time of 2.5 to 2.7 times the control value during the course of the infusion. The low dose diester compound gave a clot time of 1.4 to 1.5 times the control value. These results demonstrated the efficacy of the native and thioate analog aptamers in primates.
Example 14
Inhibition of Extracorporeal Blood Clotting By Thrombin Aptamer Anticoagulation of a hemodialysis filter was demonstrated using the 15-mer 5' GGTTGGTGTGGTTGG 3' thrombin aptamer with human blood. A bolus of 15-mer DNA was delivered to human blood at 37°C to give an aptamer concentration of lOμM. The blood was contained in an extracorporeal hemodialysis circuit (Travenol, Model No. CA-90) . Pressure proximal to the hemodialysis filter was monitored to determine the time after administration of aptamer that coagulation occurred. Blood coagulation was marked by a pressure increase from about 50 mm Hg observed with uncoagulated blood (blood flow rate 200 mL/min) to pressure of at least 400 mm Hg. Using citrated whole blood (recalcified at time zero) , coagulation occurred at about 9 minutes after fresh blood was placed in the hemodialysis unit and circulation was begun. (In a repeat of this control experiment, coagulation occurred at 11 minutes.) A heparin control (1 U/mL) gave sustained anticoagulation
until the experiment was terminated at 80 minutes after start of circulation in the unit. Blood coagulation occurred at 51 minutes in one trial with the 15-mer. In a second trial, coagulation did not occur during the 80 minute course of the experiment.
Thus, methods for obtaining aptamers that specifically bind thrombin are described, as well as the therapeutic utility of these aptamers and the use of the aptamers in the detection and isolation of thrombin. .Although preferred embodiments of the subject invention have been described in some detail, it is understood that obvious variations can be made without departing from the spirit and scope of the appended claims.
Claims
1. An aptamer containing a binding region capable of binding specifically to thrombin.
2. The aptamer of claim 1 wherein the aptamer is selected from the group consisting of single-stranded RNA, single-stranded DNA, double-stranded DNA and chemical modifications thereof.
3. The aptamer of claim 2 wherein the aptamer is single-stranded RNA.
4. The aptamer of claim 2 wherein the aptamer is single-stranded DNA.
5. The aptamer of claim 2 wherein the aptamer is double-stranded DNA.
6. An aptamer containing at least one binding region capable of binding specifically to thrombin with ;
_g dissociation constant (Kd) of less than 100 x 10 .
7. The aptamer of claim 6 containing at least one binding region capable of binding specifically to thrombin with a dissociation constant (Kd) of less than 30 x 10"9.
8. An aptamer containing at least one binding region capable of binding specifically to thrombin wherein said binding region contains less than 16 nucleotide residues.
9. The aptamer of claim 8 wherein said binding region contains more than 5 and less than 16 nucleotide residues.
10. The aptamer of claim 1 wherein the aptamer contains at least one modified base, sugar, or linking group.
11. The aptamer of claim 10 wherein the aptamer contains at least one linking group wherein P(0)0 is replaced by P(0)S, P(S)S, P(0)NR2, P(0)R, P(0)OR', CO or CH2, wherein each R or R' is independently H or substituted or unsubstituted alkyl (1-20C) optionally containing an ether (-0-) linkage, aryl, alkenyl, cycloalkyl, cycloalkenyl or aralkyl; or the aptamer contains at least one linking group attached to an adjacent nucleotide through S or N; or the aptamer contains at least one analogous form of purine or pyrimidine, or at least one abasic site.
12. The aptamer of claim 11 which is a single- stranded DNA.
13. The aptamer of claim 11 which contains at least one linking group wherein P(0)0 is replaced by P(0)S, and wherein said linking group is attached to each adjacent nucleotide through 0.
14. The aptamer of claim 11 which contains at least one linking group wherein P(0)0 is replaced by P(0)NH(CH2CH2OCH3) , and wherein said linking group is attached to each adjacent nucleotide through 0.
15. The aptamer of claim 11 which contains at least one linking group wherein P(0)0 is replaced by CH-,, and wherein said linking group is attached to each adjacent nucleotide through 0.
16. The aptamer of claim 11 wherein the aptamer is single- or double-stranded DNA and contains at least one uracil (dU) base substituted for thymine.
17. The aptamer of claim 11 containing at least one 5-pentynyluracil base substituted for thymine.
18. The aptamer of claim 11 containing at least one abasic site.
19. An aptamer capable of binding specifically to thrombin wherein the aptamer contains at least one modified or analogous sugar.
20. The aptamer of claim 19 wherein the at least one modified or analogous sugar is a furanose sugar.
21. The aptamer of claim 20 wherein the furanose sugar is a 2'-modified furanose sugar.
22. The aptamer of claim 21 wherein the 2'- modified furanose sugar is a 2' -0-alkyl-, 2'-S-alkyl-, or 2'-0-halo furanose sugar.
23. An aptamer capable of binding specifically to thrombin wherein the aptamer contains a 3'- or 5' - phosphorylated hydroxyl group.
24. The aptamer of claims 1-23 wherein said binding region comprises the sequence GGXTGG, wherein X is T, A, U, dU or G.
25. The aptamer of claim 24 wherein said nucleotide sequence has the formula GGTTGG.
26. The aptamer of claim 24 wherein said thrombin binding region comprises the sequence GGXTGG(N) -GGXTGG or a fragment thereof, wherein N is G,
A, C, U, dU or T, and z is an integer from 2 to 5.
27. The aptamer of claim 26 wherein said sequence has the formula GGTTGGTGTGGTTGG.
28. The aptamer of claim 27 having the formula GGTTGGTGTGGTTGG#G*T wherein * denotes an MEA linkage.
29. The aptamer of claim 27 having the formula GGTTGGTGTGGTT*G*G wherein * denotes a thioate linkage.
30. The aptamer of claim 27 having the formula
G G T T G G T G T G G T T G G wherein denotes a thioate linkage .
31. The aptamer of claim 27 having the formula GGTTGG(dU)G(dU)GGTTGG.
32. The aptamer of claim 27 having the formula GG(dU)TGGTGTGG(dU)TGG.
33. The aptamer of claim 27 having the formula GGTTGGTGTGGTU'GG wherein U' denotes 5-pentynyluracil.
34. The aptamer of claim 27 having the formula GGTYGGTGTGGTYGG wherein each Y is selected from the group consisting of thymine and 5-propynyluracil.
35. The aptamer of claim 27 having the formula
GGTYGGZGTGGTYGG wherein each Y is selected from the group consisting of thymine and 5-propynyluracil, and Z is an abasic site.
36. The aptamer of claim 27 having the formula
GGTY'GG(dU)G(dU)GGTY'GG wherein Y' is 5-propynyluracil.
37. The aptamer of claims 1-25 which contains a binding region of less than 16 nucleotide residues.
38. The aptamer of claims 1-23 which contains a binding region of less than 10 nucleotide residues.
39. The aptamer of claims 1-33 which contains 6-100 nucleotide residues.
40. The aptamer of claim 39 which contains 6- 50 nucleotide residues.
41. The aptamer of claims 1-40 wherein said aptamer is capable of binding specifically to thrombin at physiological conditions.
42. The aptamer of claims 1-40 wherein said aptamer binds to thrombin with a Kd of less than 100 x
10"9.
43. The aptamer of claim 42 wherein said aptamer binds to thrombin with a Kd of less than 100 x 10 -9 at physiological conditions.
44. The aptamer of claims 1-43 wherein the Kd with respect to the aptamer and thrombin is less by a factor of at least 5, as compared to the Kd for said aptamer and other molecules.
45. The aptamer of claims 1-44 which is a secondary aptamer.
46. A method for obtaining an aptamer containing at least one binding region that specifically binds thrombin, which method comprises:
(a) incubating thrombin with a mixture of oligonucleotides under conditions wherein complexation occurs with some, but not all, members of the mixture to form oligonucleotide-thrombin complexes;
(b) separating the oligonucleotide-thrombin complexes from uncomplexed oligonucleotide;
(c) recovering and amplifying the complexed oligonucleotide from said complexes; and
(d) optionally determining the sequence of the recovered oligonucleotide.
47. The method of claim 46 wherein said aptamer is a single-stranded DNA, or wherein said aptamer contains at least one binding region capable of binding specifically to thrombin with a dissociation constant (Kd) of less than 30 x 10"9, or wherein said aptamer contains at least one binding region capable of binding specifically to thrombin wherein the Kd with respect to the aptamer and thrombin is less by a factor of at least 10, as compared to the Kd for said aptamer and other molecules, or wherein said aptamer contains at least one binding region capable of binding specifically to thrombin wherein said binding region contains less than 16 nucleotide residues.
48. The method of claim 47 wherein said mixture of oligonucleotides contains at least one modified oligonucleotide.
49. The method of claim 47 wherein said amplifying is conducted using at least one modified nucleotide.
50. The method of claims 47-49 wherein said mixture of oligonucleotides contains at least one randomized-sequence region.
51. The method of claims 47-50 which further includes repeating steps (a) - (c) using the recovered and amplified complexed oligonucleotides resulting from step (c) in succeeding step (a) .
52. The method of claims 47-51 wherein the binding affinity of an oligonucleotide mixture for thrombin is at least 50-fold less than the binding affinity of the aptamer for thrombin.
53. An aptamer prepared by the method of claims 47-52.
54. A method to obtain a secondary aptamer fo: thrombin which method comprises:
(a) incubating thrombin with a mixture of oligonucleotide sequences under conditions wherein complexation occurs with some, but not all, members of the mixture to form oligonucleotide-thrombin complexes;
(b) separating the oligonucleotide-thrombin complexes from uncomplexed oligonucleotides; (c) recovering and amplifying the complexed oligonucleotides from said complexes;
(d) optionally repeating steps (a) - (c) with the recovered oligonucleotides of step (c) ;
(e) determining the sequences of the recovered oligonucleotides;
(f) determining a consensus sequence included in the recovered oligonucleotides; and
(g) synthesizing a secondary aptamer which comprises the consensus sequence.
55. A secondary aptamer prepared by the method of claim 54.
56. A complex formed by thrombin and the aptamer of claims 1-45, 53, or 55.
57. A method for obtaining an aptamer containing at least one binding region that specifically binds thrombin, which method comprises: (a) incubating thrombin with a mixture of oligonucleotides under conditions wherein complexation occurs with some, but not all, members of the mixture to form oligonucleotide-thrombin complexes;
(b) separating the oligonucleotide-thrombin complexes from uncomplexed oligonucleotide;
(c) recovering and amplifying the complexed oligonucleotide from said complexes; and
(d) optionally determining the sequence of the recovered oligonucleotide, wherein the dissociation constant (Kd) with respect to said thrombin and mixture of oligonucleotides is > 1 μM, or wherein the Kd with respect to the aptamer and 5 said thrombin is less by a factor of at least 50 as compared to the Kd for said thrombin and said mixture of oligonucleotides; or wherein steps (a) and (b) are conducted under physiological conditions, or 10 wherein said mixture of oligonucleotides consists of single-stranded DNA.
58. The method of claim 57 wherein said mixture of oligonucleotides contains at least one
15 modified oligonucleotide.
59. The method of claim 57 wherein said amplifying is conducted using at least one modified nucleotide.
20
60. The method of claim 59 wherein said at least one modified nucleotide is a 5-alkyl-2'-deoxy- pyrimidine.
25 61. The method of claim 60 wherein said 5- alkyl-2' - eoxypyrimidine is selected from the group consisting of 5-methylcytosine, 5-pentynyl-deoxyuracil, and 5-propynyl-deoxyuracil.
3C 62. The method of claims 57-61 wherein said mixture of oligonucleotides contains at least one randomized-sequence region.
63. The method of claims 57-62 which further
Ό. includes repeating steps (a) - (c) using the recovered and amplified complexed oligonucleotides resulting from step (c) in succeeding step (a) .
64. An aptamer prepared by the method of claims 57-63.
65. A method to detect the presence or absence of thrombin, which method comprises contacting a sample suspected of containing thrombin with the aptamer of claims 1-45 under conditions wherein a complex between thrombin and the aptamer is formed, and detecting the presence or absence of said complex.
66. A method to purify thrombin, which method comprises contacting a sample containing thrombin with the aptamer of claims 1-45 attached to solid support under conditions wherein thrombin is bound to the aptamer coupled to solid support; washing unbound components of the sample; and recovering thrombin from said solid support.
67. A pharmaceutical composition for medical use comprising the aptamer of claims 1-45 in admixture with a physiologically acceptable excipient.
68. A composition for diagnostic use which comprises the aptamer of claims 1-45.
69. The aptamer of claims 1-45 coupled to an auxiliary substance.
70. The aptamer of claim 69 wherein said auxiliary substance is selected from the group consisting of a drug, a toxin, a solid support, and specific binding reagent, a label, a radioisotope, or a contrast agent.
71. The aptamer of claim 70 wherein said auxiliary substance is a radioisotope selected from the group consisting of 131I, 99aTc, 90Y, 111In and 1 3I.
72. A method to obtain an aptamer containing a binding region which specifically binds thrombin which comprises:
(a) incubating thrombin reversibly coupled to a support with a mixture of oligonucleotide sequences under conditions wherein the coupled thrombin complexes with some, but not all, members of the mixture to form support-bound oligonucleotide complexes;
(b) decoupling and recovering the oligonucleotide-thrombin complex from the support to obtain free aptamer-thrombin complexes;
(c) recovering and amplifying the complexed oligonucleotides from the free oligonucleotide-thrombin complexes to obtain a population of aptamers;
(d) optionally repeating steps (a) - (c) using as said mixture the recovered population of aptamers of step (c) ; and (e) optionally determining the sequence of the recovered aptamers.
73. The method of claim 72 wherein the support is a lectin support.
74. The method of claim 73 wherein in step (b) , decoupling is accomplished by adding a monosaccharide.
75. The method of claim 74 wherein the monosaccharide is selected from the group consisting of α-methyl-mannoside, N-acetylglucosamine, glucose, N- acetylgalactosa ine and galactose.
76. The method of claim 75 wherein the support is a concanavalin A column.
77. A composition for use in binding or inhibiting thrombin which composition comprises an aptamer as described in claims 1-45.
77. A composition for use in inhibiting clotting or coagulation in a patient's blood which composition comprises an aptamer as described in claims 1-45.
78. A composition for use in inhibiting or reducing restenosis, which composition comprises an aptamer as described in claims 1-45.
79. A composition for use in treating a patient's blood ex corpore to inhibit clot formation, which composition comprises an aptamer as described in claims 1-45.
80. A method to prevent coagulation during cardiopulmonary bypass surgery, which method comprises contacting blood with an aptamer as described in claims 1-45.
81. In a method to inhibit clot formation which comprises contacting blood with a fibrinolytic agent, the improvement which comprises: contacting said blood with an aptamer as described in claims 1-45.
Applications Claiming Priority (10)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| US65884991A | 1991-02-21 | 1991-02-21 | |
| US65998191A | 1991-02-21 | 1991-02-21 | |
| US658,849 | 1991-02-21 | ||
| US659,981 | 1991-02-21 | ||
| US74521591A | 1991-08-14 | 1991-08-14 | |
| US74487091A | 1991-08-14 | 1991-08-14 | |
| US744,870 | 1991-08-14 | ||
| US745,215 | 1991-08-14 | ||
| US78792191A | 1991-11-06 | 1991-11-06 | |
| US787,921 | 1991-11-06 |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| WO1992014842A1 true WO1992014842A1 (en) | 1992-09-03 |
Family
ID=27542045
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/US1992/001367 WO1992014842A1 (en) | 1991-02-21 | 1992-02-21 | Aptamers specific for thrombin and methods of use |
Country Status (4)
| Country | Link |
|---|---|
| AU (1) | AU1456092A (en) |
| IE (1) | IE920561A1 (en) |
| IL (1) | IL101038A0 (en) |
| WO (1) | WO1992014842A1 (en) |
Cited By (42)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO1993012135A1 (en) | 1991-12-12 | 1993-06-24 | Gilead Sciences, Inc. | Nuclease stable and binding competent oligomers and methods for their use |
| US5639595A (en) * | 1990-05-01 | 1997-06-17 | Isis Phamaceuticals, Inc. | Identification of novel drugs and reagents |
| EP0786469A2 (en) * | 1990-06-11 | 1997-07-30 | NeXstar Pharmaceuticals, Inc. | Nucleic acid ligands |
| EP0824540A1 (en) * | 1995-05-03 | 1998-02-25 | NeXstar Pharmaceuticals, Inc. | Nucleic acid ligands of tissue target |
| EP0968226A1 (en) * | 1996-12-27 | 2000-01-05 | ICN Pharmaceuticals, Inc. | G-rich oligo aptamers and methods of modulating an immune response |
| US6206860B1 (en) | 1993-07-28 | 2001-03-27 | Frank M. Richmond | Spikeless connection and drip chamber with valve |
| EP1143980A1 (en) * | 1998-10-26 | 2001-10-17 | Board of Regents, The University of Texas System | Thio-modified aptamer synthetic methods and compositions |
| WO2002096926A1 (en) * | 2001-05-25 | 2002-12-05 | Duke University | Modulators of pharmacological agents |
| JP2003517041A (en) * | 1999-12-13 | 2003-05-20 | バイオニケ ライフ サイエンシーズ インコーポレイテッド | Therapeutically useful synthetic oligonucleotides |
| US6780850B1 (en) | 1999-06-22 | 2004-08-24 | Triumf | Extending the lifetime of anticoagulant oligodeoxynucleotide aptamers in blood |
| EP1489171A1 (en) * | 2002-03-19 | 2004-12-22 | Fujitsu Limited | Functional molecule and process for producing the same |
| EP1534301A2 (en) * | 2001-11-15 | 2005-06-01 | Board Of Regents The University Of Texas System | Phosphoromonothioate and phosphorodithioate oligonucleotide aptamer chip for functional proteomics |
| EP1552002A2 (en) * | 2002-06-18 | 2005-07-13 | Archemix Corp. | Aptamer-toxin molecules and methods for using same |
| US7304041B2 (en) | 2004-04-22 | 2007-12-04 | Regado Biosciences, Inc. | Modulators of coagulation factors |
| US7312325B2 (en) | 2000-09-26 | 2007-12-25 | Duke University | RNA aptamers and methods for identifying the same |
| US7338762B2 (en) | 2002-10-16 | 2008-03-04 | Board Of Regents, The University Of Texas System | Bead bound combinatorial oligonucleoside phosphorothioate and phosphorodithioate aptamer libraries |
| US7538211B2 (en) | 2004-02-12 | 2009-05-26 | Archemix Corp. | Aptamer therapeutics useful in the treatment of complement-related disorders |
| US7579450B2 (en) | 2004-04-26 | 2009-08-25 | Archemix Corp. | Nucleic acid ligands specific to immunoglobulin E and their use as atopic disease therapeutics |
| US7803931B2 (en) | 2004-02-12 | 2010-09-28 | Archemix Corp. | Aptamer therapeutics useful in the treatment of complement-related disorders |
| US7855054B2 (en) | 2007-01-16 | 2010-12-21 | Somalogic, Inc. | Multiplexed analyses of test samples |
| US7910523B2 (en) | 2003-05-23 | 2011-03-22 | Board Of Regents, The University Of Texas System | Structure based and combinatorially selected oligonucleoside phosphorothioate and phosphorodithioate aptamer targeting AP-1 transcription factors |
| US7947447B2 (en) | 2007-01-16 | 2011-05-24 | Somalogic, Inc. | Method for generating aptamers with improved off-rates |
| US7960102B2 (en) | 2002-07-25 | 2011-06-14 | Archemix Corp. | Regulated aptamer therapeutics |
| US7964356B2 (en) | 2007-01-16 | 2011-06-21 | Somalogic, Inc. | Method for generating aptamers with improved off-rates |
| US7964572B2 (en) | 1996-02-01 | 2011-06-21 | Gilead Sciences, Inc. | High affinity nucleic acid ligands of complement system proteins |
| US7998939B2 (en) | 2005-08-26 | 2011-08-16 | Archemix Corporation | Aptamers that bind thrombin with high affinity |
| US8071288B2 (en) | 1997-12-15 | 2011-12-06 | Somalogic, Inc. | Methods and reagents for detecting target binding by nucleic acid ligands |
| WO2013073602A1 (en) | 2011-11-18 | 2013-05-23 | 独立行政法人理化学研究所 | Nucleic acid fragment binding to target protein |
| US8598140B2 (en) | 2010-04-12 | 2013-12-03 | Somalogic, Inc. | Aptamers to β-NGF and their use in treating β-NGF mediated diseases and disorders |
| US8703416B2 (en) | 2008-07-17 | 2014-04-22 | Somalogic, Inc. | Method for purification and identification of sperm cells |
| US8853376B2 (en) | 2002-11-21 | 2014-10-07 | Archemix Llc | Stabilized aptamers to platelet derived growth factor and their use as oncology therapeutics |
| US8945830B2 (en) | 1997-12-15 | 2015-02-03 | Somalogic, Inc. | Multiplexed analyses of test samples |
| US8975026B2 (en) | 2007-01-16 | 2015-03-10 | Somalogic, Inc. | Method for generating aptamers with improved off-rates |
| US9303262B2 (en) | 2002-09-17 | 2016-04-05 | Archemix Llc | Methods for identifying aptamer regulators |
| US9365855B2 (en) | 2010-03-03 | 2016-06-14 | Somalogic, Inc. | Aptamers to 4-1BB and their use in treating diseases and disorders |
| US9404919B2 (en) | 2007-01-16 | 2016-08-02 | Somalogic, Inc. | Multiplexed analyses of test samples |
| EP3085786A3 (en) * | 2011-03-07 | 2016-11-30 | Charité - Universitätsmedizin Berlin | Use of aptamers in therapy and/or diagnosis of autoimmune diseases |
| WO2017100305A3 (en) * | 2015-12-07 | 2017-07-20 | Opi Vi - Ip Holdco Llc | Composition of antibody construct-agonist conjugates and methods of use thereof |
| US9910034B2 (en) | 2007-11-06 | 2018-03-06 | Ambergen, Inc. | Methods and compositions for phototransfer |
| WO2018144955A1 (en) * | 2017-02-02 | 2018-08-09 | Silverback Therapeutics, Inc. | Construct-peptide compositions and methods of use thereof |
| US10100316B2 (en) | 2002-11-21 | 2018-10-16 | Archemix Llc | Aptamers comprising CPG motifs |
| CN111440235A (en) * | 2020-04-16 | 2020-07-24 | 成都中医药大学 | Probe for capturing hirudin polypeptide and application thereof |
Citations (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US4647529A (en) * | 1984-06-01 | 1987-03-03 | Rodland Karin D | Hybridization method of detecting nucleic acid sequences with probe containing thionucleotide |
| US4748156A (en) * | 1986-01-21 | 1988-05-31 | Kowa Co., Ltd. | Thrombin-binding substance and process for preparing the same |
-
1992
- 1992-02-21 AU AU14560/92A patent/AU1456092A/en not_active Abandoned
- 1992-02-21 WO PCT/US1992/001367 patent/WO1992014842A1/en active Application Filing
- 1992-02-21 IL IL101038A patent/IL101038A0/en unknown
- 1992-02-21 IE IE056192A patent/IE920561A1/en unknown
Patent Citations (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US4647529A (en) * | 1984-06-01 | 1987-03-03 | Rodland Karin D | Hybridization method of detecting nucleic acid sequences with probe containing thionucleotide |
| US4748156A (en) * | 1986-01-21 | 1988-05-31 | Kowa Co., Ltd. | Thrombin-binding substance and process for preparing the same |
Non-Patent Citations (7)
| Title |
|---|
| KIRK-OTHMER, ENCYCLOPEIDA OF CHEMICAL TECHNOLOGY, Third Edition, Volume 6, published 1979 by JOHN-WILEY AND SONS (NY), pages 35-54. * |
| NATURE, Volume 346, issued 30 August 1990, ELLINGTON et al., "In vitro Selection of RNA Molecules that Bind Specific Ligands", pages 818-822. * |
| NUCLEIC ACIDS RESEARCH, Volume 17, Number 10, issued 1989, KINZLER et al., "Whole Genome PCR: Application to the Identification of Sequences Bound by Gene Regulatory Proteins", pages 3645-3653. * |
| NUCLEIC ACIDS RESEARCH, Volume 18, Number 11, issued 1990, THIESEN et al., "Target Detection Assay (TDA): A Versatile Procedure to Determine DNA Binding Sites as Demonstrated on SP1 Protein", pages 3203-3208. * |
| PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES, Vol. 82, issued November 1985, HUYNH-DINH et al., "Modified Oligonucleotides as Alternatives to the Synthesis of Mixed Probes for the Screening of cDNA Libraries", pages 7510-7514. * |
| SCIENCE, Volume 249, issued 03 August 1990, TUERK et al., "Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase", pages 505-510. * |
| SCIENCE, Volume 250, issued 23 November 1990, BLACKWELL et al., "Differences and Similarities in DNA-Binding Preferences of MyoD and E2A Protein Complexes Revealed by Binding Site Selection", pages 1104-1110. * |
Cited By (93)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US5639595A (en) * | 1990-05-01 | 1997-06-17 | Isis Phamaceuticals, Inc. | Identification of novel drugs and reagents |
| EP0786469A2 (en) * | 1990-06-11 | 1997-07-30 | NeXstar Pharmaceuticals, Inc. | Nucleic acid ligands |
| EP0786469A3 (en) * | 1990-06-11 | 1997-08-06 | NeXstar Pharmaceuticals, Inc. | Nucleic acid ligands |
| US6933116B2 (en) | 1990-06-11 | 2005-08-23 | Gilead Sciences, Inc. | Nucleic acid ligand binding site identification |
| US5843653A (en) * | 1990-06-11 | 1998-12-01 | Nexstar Pharmaceuticals, Inc. | Method for detecting a target molecule in a sample using a nucleic acid ligand |
| EP1493825A3 (en) * | 1990-06-11 | 2005-02-09 | Gilead Sciences, Inc. | Method for producing nucleic acid ligands |
| US6110900A (en) * | 1990-06-11 | 2000-08-29 | Nexstar Pharmaceuticals, Inc. | Nucleic acid ligands |
| EP1493825A2 (en) * | 1990-06-11 | 2005-01-05 | Gilead Sciences, Inc. | Method for producing nucleic acid ligands |
| EP1695978A1 (en) * | 1990-06-11 | 2006-08-30 | Gilead Sciences, Inc. | Nucleic acid ligands |
| US6331398B1 (en) | 1990-06-11 | 2001-12-18 | Gilead Sciences, Inc. | Nucleic acid ligands |
| WO1993012135A1 (en) | 1991-12-12 | 1993-06-24 | Gilead Sciences, Inc. | Nuclease stable and binding competent oligomers and methods for their use |
| US6206860B1 (en) | 1993-07-28 | 2001-03-27 | Frank M. Richmond | Spikeless connection and drip chamber with valve |
| EP2000534A3 (en) * | 1995-05-03 | 2009-09-02 | Gilead Sciences, Inc. | Nucleic acid ligands of tissue target related applications |
| EP0824540A4 (en) * | 1995-05-03 | 2003-04-23 | Nexstar Pharmaceuticals Inc | Nucleic acid ligands of tissue target |
| EP1564290A2 (en) * | 1995-05-03 | 2005-08-17 | Gilead Sciences, Inc. | Nucleic acid ligands of tissue targets |
| EP1564290A3 (en) * | 1995-05-03 | 2006-02-01 | Gilead Sciences, Inc. | Nucleic acid ligands of tissue targets |
| EP0824540A1 (en) * | 1995-05-03 | 1998-02-25 | NeXstar Pharmaceuticals, Inc. | Nucleic acid ligands of tissue target |
| US7964572B2 (en) | 1996-02-01 | 2011-06-21 | Gilead Sciences, Inc. | High affinity nucleic acid ligands of complement system proteins |
| EP0968226A4 (en) * | 1996-12-27 | 2001-02-14 | Icn Pharmaceuticals | G-rich oligo aptamers and methods of modulating an immune response |
| EP0968226A1 (en) * | 1996-12-27 | 2000-01-05 | ICN Pharmaceuticals, Inc. | G-rich oligo aptamers and methods of modulating an immune response |
| US8945830B2 (en) | 1997-12-15 | 2015-02-03 | Somalogic, Inc. | Multiplexed analyses of test samples |
| US8071288B2 (en) | 1997-12-15 | 2011-12-06 | Somalogic, Inc. | Methods and reagents for detecting target binding by nucleic acid ligands |
| US7179894B2 (en) | 1998-10-26 | 2007-02-20 | Board Of Regents, The University Of Texas System | Combinatorial selection of oligonucleotide aptamers |
| EP1143980A1 (en) * | 1998-10-26 | 2001-10-17 | Board of Regents, The University of Texas System | Thio-modified aptamer synthetic methods and compositions |
| US6867289B1 (en) | 1998-10-26 | 2005-03-15 | Board Of Regents, The University Of Texas Systems | Thio-modified aptamer synthetic methods and compositions |
| EP1143980A4 (en) * | 1998-10-26 | 2003-07-02 | Regents Board Of | Thio-modified aptamer synthetic methods and compositions |
| US6780850B1 (en) | 1999-06-22 | 2004-08-24 | Triumf | Extending the lifetime of anticoagulant oligodeoxynucleotide aptamers in blood |
| JP2003517041A (en) * | 1999-12-13 | 2003-05-20 | バイオニケ ライフ サイエンシーズ インコーポレイテッド | Therapeutically useful synthetic oligonucleotides |
| JP4772245B2 (en) * | 1999-12-13 | 2011-09-14 | バイオニケ ライフ サイエンシーズ インコーポレイテッド | Therapeutically useful synthetic oligonucleotides |
| US7858591B2 (en) | 2000-09-26 | 2010-12-28 | Duke University | RNA aptamers and methods for identifying the same |
| US7812001B2 (en) | 2000-09-26 | 2010-10-12 | Duke University | RNA aptamers and methods for identifying the same |
| EP2322648A1 (en) * | 2000-09-26 | 2011-05-18 | Duke University | RNA aptamers and methods for identifying the same |
| US7312325B2 (en) | 2000-09-26 | 2007-12-25 | Duke University | RNA aptamers and methods for identifying the same |
| US7776837B2 (en) | 2000-09-26 | 2010-08-17 | Duke University | RNA aptamers and methods for identifying the same |
| US7776836B2 (en) | 2000-09-26 | 2010-08-17 | Duke University | RNA aptamers and methods for identifying the same |
| US7741307B2 (en) | 2000-09-26 | 2010-06-22 | Duke University | RNA aptamers and methods for identifying the same |
| US8143233B2 (en) | 2000-09-26 | 2012-03-27 | Duke University | RNA aptamers and methods for identifying the same |
| US8586524B2 (en) | 2001-05-25 | 2013-11-19 | Duke University | Modulators of pharmacological agents |
| US7300922B2 (en) | 2001-05-25 | 2007-11-27 | Duke University | Modulators of pharmacological agents |
| SG157961A1 (en) * | 2001-05-25 | 2010-01-29 | Univ Duke | Modulators of pharmacological agents |
| WO2002096926A1 (en) * | 2001-05-25 | 2002-12-05 | Duke University | Modulators of pharmacological agents |
| US8283330B2 (en) | 2001-05-25 | 2012-10-09 | Duke University | Modulators of pharmacological agents |
| EP1534301A2 (en) * | 2001-11-15 | 2005-06-01 | Board Of Regents The University Of Texas System | Phosphoromonothioate and phosphorodithioate oligonucleotide aptamer chip for functional proteomics |
| EP1534301A4 (en) * | 2001-11-15 | 2007-06-20 | Univ Texas | Phosphoromonothioate and phosphorodithioate oligonucleotide aptamer chip for functional proteomics |
| US7517646B2 (en) | 2002-03-19 | 2009-04-14 | Fujitsu Limited | Functional molecule and process for producing the same |
| EP1489171A4 (en) * | 2002-03-19 | 2008-05-07 | Fujitsu Ltd | Functional molecule and process for producing the same |
| EP1489171A1 (en) * | 2002-03-19 | 2004-12-22 | Fujitsu Limited | Functional molecule and process for producing the same |
| EP1552002A2 (en) * | 2002-06-18 | 2005-07-13 | Archemix Corp. | Aptamer-toxin molecules and methods for using same |
| EP1552002A4 (en) * | 2002-06-18 | 2006-02-08 | Archemix Corp | Aptamer-toxin molecules and methods for using same |
| US7960102B2 (en) | 2002-07-25 | 2011-06-14 | Archemix Corp. | Regulated aptamer therapeutics |
| US9303262B2 (en) | 2002-09-17 | 2016-04-05 | Archemix Llc | Methods for identifying aptamer regulators |
| US7338762B2 (en) | 2002-10-16 | 2008-03-04 | Board Of Regents, The University Of Texas System | Bead bound combinatorial oligonucleoside phosphorothioate and phosphorodithioate aptamer libraries |
| US10100316B2 (en) | 2002-11-21 | 2018-10-16 | Archemix Llc | Aptamers comprising CPG motifs |
| US8853376B2 (en) | 2002-11-21 | 2014-10-07 | Archemix Llc | Stabilized aptamers to platelet derived growth factor and their use as oncology therapeutics |
| US7910523B2 (en) | 2003-05-23 | 2011-03-22 | Board Of Regents, The University Of Texas System | Structure based and combinatorially selected oligonucleoside phosphorothioate and phosphorodithioate aptamer targeting AP-1 transcription factors |
| US9567579B2 (en) | 2003-05-23 | 2017-02-14 | Board Of Regents, The University Of Texas System | Structure based and combinatorially selected oligonucleoside phosphorothioate and phosphorodithioate aptamer targeting AP-1 transcription factors |
| US7803931B2 (en) | 2004-02-12 | 2010-09-28 | Archemix Corp. | Aptamer therapeutics useful in the treatment of complement-related disorders |
| US7538211B2 (en) | 2004-02-12 | 2009-05-26 | Archemix Corp. | Aptamer therapeutics useful in the treatment of complement-related disorders |
| US7579456B2 (en) | 2004-02-12 | 2009-08-25 | Archemix Corp. | Aptamer therapeutics useful in the treatment of complement-related disorders |
| US7304041B2 (en) | 2004-04-22 | 2007-12-04 | Regado Biosciences, Inc. | Modulators of coagulation factors |
| US8389489B2 (en) | 2004-04-22 | 2013-03-05 | Regado Biosciences, Inc. | Modulators of coagulation factors |
| US7723315B2 (en) | 2004-04-22 | 2010-05-25 | Regado Biosciences, Inc. | Modulators of coagulation factors |
| US8859518B2 (en) | 2004-04-22 | 2014-10-14 | Regado Biosciences, Inc. | Modulators of coagulation factors |
| US7579450B2 (en) | 2004-04-26 | 2009-08-25 | Archemix Corp. | Nucleic acid ligands specific to immunoglobulin E and their use as atopic disease therapeutics |
| US9617546B2 (en) | 2005-02-14 | 2017-04-11 | Archemix Llc | Aptamer therapeutics useful in the treatment of complement-related disorders |
| US10947544B2 (en) | 2005-02-14 | 2021-03-16 | Archemix Llc | Aptamer therapeutics useful in the treatment of complement-related disorders |
| US11913000B2 (en) | 2005-02-14 | 2024-02-27 | Iveric Bio, Inc. | Aptamer therapeutics useful in the treatment of complement-related disorders |
| US8946184B2 (en) | 2005-02-14 | 2015-02-03 | Archemix Llc | Aptamer therapeutics useful in the treatment of complement-related disorders |
| US7998939B2 (en) | 2005-08-26 | 2011-08-16 | Archemix Corporation | Aptamers that bind thrombin with high affinity |
| US10648972B2 (en) | 2006-01-17 | 2020-05-12 | Somalogic, Inc. | Multiplexed analyses of test samples |
| US10422794B2 (en) | 2006-01-17 | 2019-09-24 | Somalogic, Inc. | Multiplexed analyses of test samples |
| US9404919B2 (en) | 2007-01-16 | 2016-08-02 | Somalogic, Inc. | Multiplexed analyses of test samples |
| US8975388B2 (en) | 2007-01-16 | 2015-03-10 | Somalogic, Inc. | Method for generating aptamers with improved off-rates |
| US7964356B2 (en) | 2007-01-16 | 2011-06-21 | Somalogic, Inc. | Method for generating aptamers with improved off-rates |
| US9382533B2 (en) | 2007-01-16 | 2016-07-05 | Somalogic, Inc. | Method for generating aptamers with improved off-rates |
| US8975026B2 (en) | 2007-01-16 | 2015-03-10 | Somalogic, Inc. | Method for generating aptamers with improved off-rates |
| US11111495B2 (en) | 2007-01-16 | 2021-09-07 | Somalogic, Inc. | Method for generating aptamers with improved off-rates |
| US7947447B2 (en) | 2007-01-16 | 2011-05-24 | Somalogic, Inc. | Method for generating aptamers with improved off-rates |
| US7855054B2 (en) | 2007-01-16 | 2010-12-21 | Somalogic, Inc. | Multiplexed analyses of test samples |
| US10316321B2 (en) | 2007-01-16 | 2019-06-11 | Somalogic Inc. | Method for generating aptamers with improved off-rates |
| US9910034B2 (en) | 2007-11-06 | 2018-03-06 | Ambergen, Inc. | Methods and compositions for phototransfer |
| US10088474B2 (en) | 2007-11-06 | 2018-10-02 | Ambergen, Inc. | Methods and compositions for phototransfer |
| US8703416B2 (en) | 2008-07-17 | 2014-04-22 | Somalogic, Inc. | Method for purification and identification of sperm cells |
| US9365855B2 (en) | 2010-03-03 | 2016-06-14 | Somalogic, Inc. | Aptamers to 4-1BB and their use in treating diseases and disorders |
| US8598140B2 (en) | 2010-04-12 | 2013-12-03 | Somalogic, Inc. | Aptamers to β-NGF and their use in treating β-NGF mediated diseases and disorders |
| EP3085786A3 (en) * | 2011-03-07 | 2016-11-30 | Charité - Universitätsmedizin Berlin | Use of aptamers in therapy and/or diagnosis of autoimmune diseases |
| US9873879B2 (en) | 2011-11-18 | 2018-01-23 | Tagcyx Biotechnologies | Nucleic acid fragment binding to target protein |
| KR20140091750A (en) | 2011-11-18 | 2014-07-22 | 탁시스 바이오 가부시키가이샤 | Nucleic acid fragment binding to target protein |
| WO2013073602A1 (en) | 2011-11-18 | 2013-05-23 | 独立行政法人理化学研究所 | Nucleic acid fragment binding to target protein |
| US9540650B2 (en) | 2011-11-18 | 2017-01-10 | Tagcyx Biotechnologies | Nucleic acid fragment binding to target protein |
| WO2017100305A3 (en) * | 2015-12-07 | 2017-07-20 | Opi Vi - Ip Holdco Llc | Composition of antibody construct-agonist conjugates and methods of use thereof |
| WO2018144955A1 (en) * | 2017-02-02 | 2018-08-09 | Silverback Therapeutics, Inc. | Construct-peptide compositions and methods of use thereof |
| CN111440235A (en) * | 2020-04-16 | 2020-07-24 | 成都中医药大学 | Probe for capturing hirudin polypeptide and application thereof |
Also Published As
| Publication number | Publication date |
|---|---|
| IE920561A1 (en) | 1992-08-26 |
| IL101038A0 (en) | 1992-11-15 |
| AU1456092A (en) | 1992-09-15 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| WO1992014842A1 (en) | Aptamers specific for thrombin and methods of use | |
| US5840867A (en) | Aptamer analogs specific for biomolecules | |
| US5582981A (en) | Method for identifying an oligonucleotide aptamer specific for a target | |
| KR970002255B1 (en) | Nucleic acid ligands | |
| US5658738A (en) | Bi-directional oligonucleotides that bind thrombin | |
| JPH06508022A (en) | Biomolecule-specific aptamers and production methods | |
| US6083696A (en) | Systematic evolution of ligands exponential enrichment: blended selex | |
| JENISON et al. | Oligonucleotide inhibitors of P-selectin-dependent neutrophil-platelet adhesion | |
| AU777043B2 (en) | Nucleic acid ligands to CD40ligand | |
| US5683867A (en) | Systematic evolution of ligands by exponential enrichment: blended SELEX | |
| CA2448567C (en) | Modulators of nucleic acid ligands | |
| AU767501B2 (en) | Tenascin-C nucleic acid ligands | |
| CA2622765A1 (en) | Focused libraries, functional profiling, laser selex and deselex | |
| EP1793006A2 (en) | Nucleic acid ligands and improved methods for producing the same | |
| WO1999055857A2 (en) | Nucleoside triphosphates and their incorporation into ribozymes | |
| AU3679595A (en) | Parallel selex | |
| CA2504633A1 (en) | Multivalent aptamer therapeutics with improved pharmacodynamic properties and methods of making and using the same | |
| AU4993097A (en) | Morphatides: novel shape and structure libraries | |
| EP3423594B1 (en) | Oligonucleotides and methods for preparing | |
| US20020022227A1 (en) | Morphatides: novel shape and structure libraries | |
| AU2012244176B2 (en) | Modulators of pharmacological agents | |
| AU2012244176B8 (en) | Modulators of pharmacological agents |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| AK | Designated states |
Kind code of ref document: A1 Designated state(s): AT AU BB BG BR CA CH CS DE DK ES FI GB HU JP KP KR LK LU MG MN MW NL NO PL RO RU SD SE US |
|
| AL | Designated countries for regional patents |
Kind code of ref document: A1 Designated state(s): AT BE BF BJ CF CG CH CI CM DE DK ES FR GA GB GN GR IT LU MC ML MR NL SE SN TD TG |
|
| DFPE | Request for preliminary examination filed prior to expiration of 19th month from priority date (pct application filed before 20040101) |
Free format text: PL |
|
| REG | Reference to national code |
Ref country code: DE Ref legal event code: 8642 |
|
| NENP | Non-entry into the national phase |
Ref country code: CA |
|
| 122 | Ep: pct application non-entry in european phase |